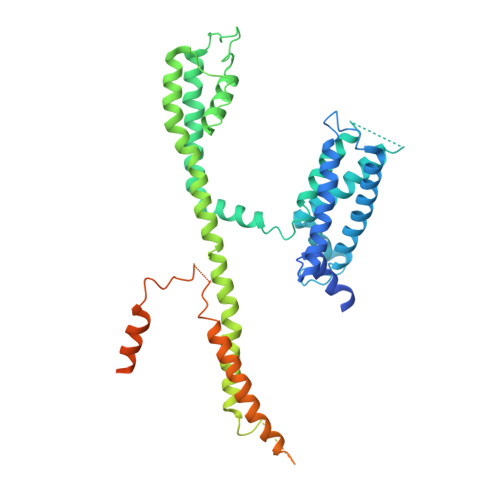
Structural Basis of Human KCNQ1 Modulation and Gating.
Sun, J., MacKinnon, R.(2020) Cell 180: 340-347.e9
- PubMed: 31883792
- DOI: https://doi.org/10.1016/j.cell.2019.12.003
- Primary Citation of Related Structures:
6UZZ, 6V00, 6V01 - PubMed Abstract:
KCNQ1, also known as Kv7.1, is a voltage-dependent K + channel that regulates gastric acid secretion, salt and glucose homeostasis, and heart rhythm. Its functional properties are regulated in a tissue-specific manner through co-assembly with beta subunits KCNE1-5. In non-excitable cells, KCNQ1 forms a complex with KCNE3, which suppresses channel closure at negative membrane voltages that otherwise would close it. Pore opening is regulated by the signaling lipid PIP2. Using cryoelectron microscopy (cryo-EM), we show that KCNE3 tucks its single-membrane-spanning helix against KCNQ1, at a location that appears to lock the voltage sensor in its depolarized conformation. Without PIP2, the pore remains closed. Upon addition, PIP2 occupies a site on KCNQ1 within the inner membrane leaflet, which triggers a large conformational change that leads to dilation of the pore's gate. It is likely that this mechanism of PIP2 activation is conserved among Kv7 channels.
- Laboratory of Molecular Neurobiology and Biophysics and Howard Hughes Medical Institute, The Rockefeller University, 1230 York Avenue, New York, NY 10065, USA.
Organizational Affiliation: