Integration of phage and yeast display platforms: A reliable and cost effective approach for binning of peptides as displayed on-phage.
Pandya, P., Sayers, R.O., Ting, J.P., Morshedian, S., Torres, C., Cudal, J.S., Zhang, K., Fitchett, J.R., Zhang, Q., Zhang, F.F., Wang, J., Durbin, J.D., Carrillo, J.J., Espada, A., Broughton, H., Qian, Y., Afshar, S.(2020) PLoS One 15: e0233961-e0233961
- PubMed: 32479512
- DOI: https://doi.org/10.1371/journal.pone.0233961
- Primary Citation of Related Structures:
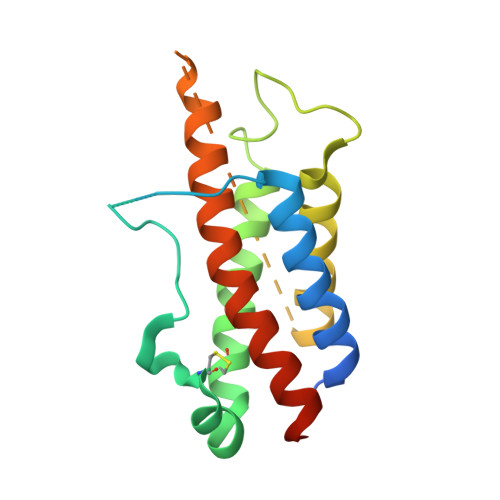
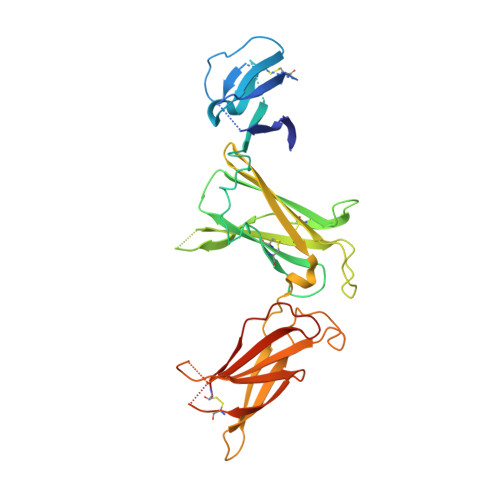
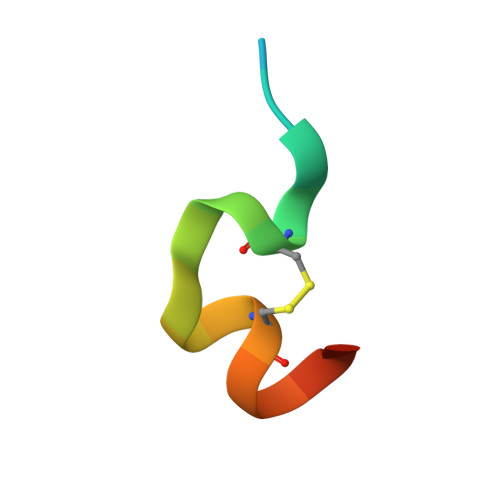
6UIB - PubMed Abstract:
Hundreds of target specific peptides are routinely discovered by peptide display platforms. However, due to the high cost of peptide synthesis only a limited number of peptides are chemically made for further analysis. Here we describe an accurate and cost effective method to bin peptides on-phage based on binding region(s), without any requirement for peptide or protein synthesis. This approach, which integrates phage and yeast display platforms, requires display of target and its alanine variants on yeast. Flow cytometry was used to detect binding of peptides on-phage to the target on yeast. Once hits were identified, they were synthesized to confirm their binding region(s) by HDX (Hydrogen deuterium exchange) and crystallography. Moreover, we have successfully shown that this approach can be implemented as part of a panning process to deplete non-functional peptides. This technique can be applied to any target that can be successfully displayed on yeast; it narrows down the number of peptides requiring synthesis; and its utilization during selection results in enrichment of peptide population against defined binding regions on the target.
- Department of Protein Engineering, Eli Lilly Biotechnology Center, San Diego, California, United States of America.
Organizational Affiliation: