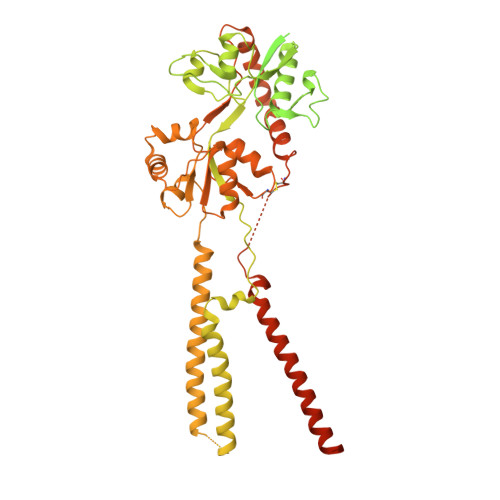
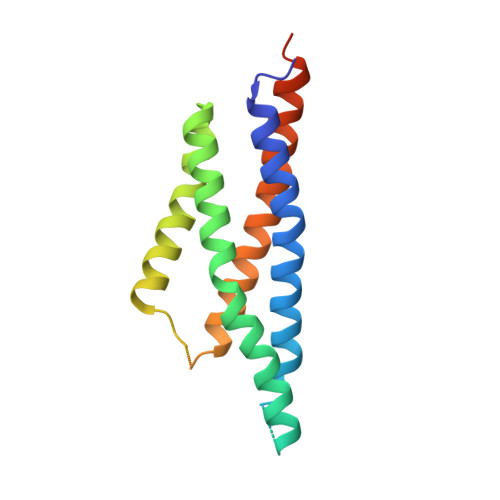
Structures of the AMPA receptor in complex with its auxiliary subunit cornichon.
Nakagawa, T.(2019) Science 366: 1259-1263
- PubMed: 31806817
- DOI: https://doi.org/10.1126/science.aay2783
- Primary Citation of Related Structures:
6PEQ, 6U5S, 6U6I, 6UCB, 6UD4, 6UD8 - PubMed Abstract:
In the brain, AMPA-type glutamate receptors (AMPARs) form complexes with their auxiliary subunits and mediate the majority of fast excitatory neurotransmission. Signals transduced by these complexes are critical for synaptic plasticity, learning, and memory. The two major categories of AMPAR auxiliary subunits are transmembrane AMPAR regulatory proteins (TARPs) and cornichon homologs (CNIHs); these subunits share little homology and play distinct roles in controlling ion channel gating and trafficking of AMPAR. Here, I report high-resolution cryo-electron microscopy structures of AMPAR in complex with CNIH3. Contrary to its predicted membrane topology, CNIH3 lacks an extracellular domain and instead contains four membrane-spanning helices. The protein-protein interaction interface that dictates channel modulation and the lipids surrounding the complex are revealed. These structures provide insights into the molecular mechanism for ion channel modulation and assembly of AMPAR/CNIH3 complexes.
- Department of Molecular Physiology and Biophysics, Center for Structural Biology, and Vanderbilt Brain Institute, Vanderbilt University School of Medicine, Nashville, TN 37232, USA.
Organizational Affiliation: