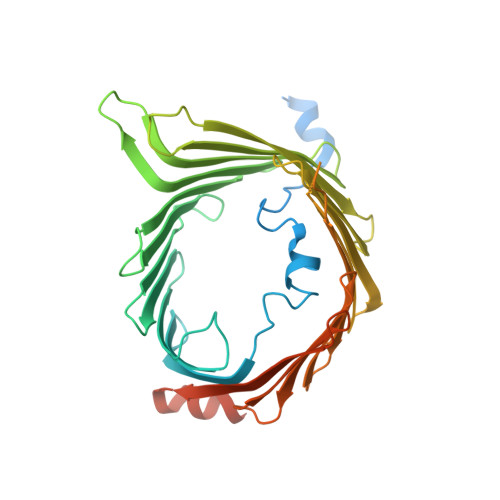
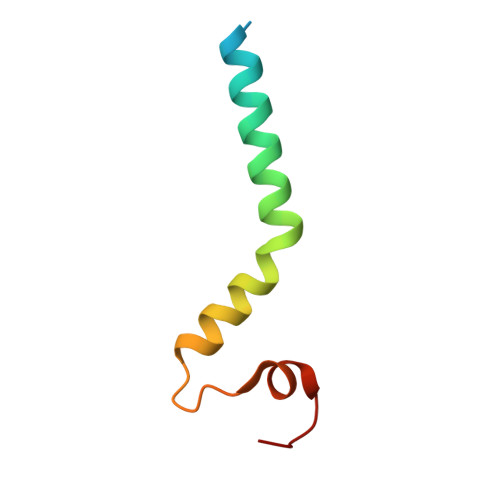
Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution.
Tucker, K., Park, E.(2019) Nat Struct Mol Biol 26: 1158-1166
- PubMed: 31740857
- DOI: https://doi.org/10.1038/s41594-019-0339-2
- Primary Citation of Related Structures:
6UCU, 6UCV - PubMed Abstract:
Nearly all mitochondrial proteins are encoded by the nuclear genome and imported into mitochondria after synthesis on cytosolic ribosomes. These precursor proteins are translocated into mitochondria by the TOM complex, a protein-conducting channel in the mitochondrial outer membrane. We have determined high-resolution cryo-EM structures of the core TOM complex from Saccharomyces cerevisiae in dimeric and tetrameric forms. Dimeric TOM consists of two copies each of five proteins arranged in two-fold symmetry: pore-forming β-barrel protein Tom40 and four auxiliary α-helical transmembrane proteins. The pore of each Tom40 has an overall negatively charged inner surface attributed to multiple functionally important acidic patches. The tetrameric complex is essentially a dimer of dimeric TOM, which may be capable of forming higher-order oligomers. Our study reveals the detailed molecular organization of the TOM complex and provides new insights about the mechanism of protein translocation into mitochondria.
- Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, CA, USA.
Organizational Affiliation: