Crystal structure of human PACRG in complex with MEIG1
Khan, N., Pelletier, D., Veyron, S., Croteau, N., Ichikawa, M., Black, C., Khalifa, A.A.Z., Chaaban, S., Kurinov, I., Brouhard, G., Bui, K.H., Trempe, J.F.(2019) bioRxiv
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Currently 6UCC does not have a validation slider image.
(2019) bioRxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Parkin coregulated gene protein | 257 | Homo sapiens | Mutation(s): 0 Gene Names: PACRG, GLUP | 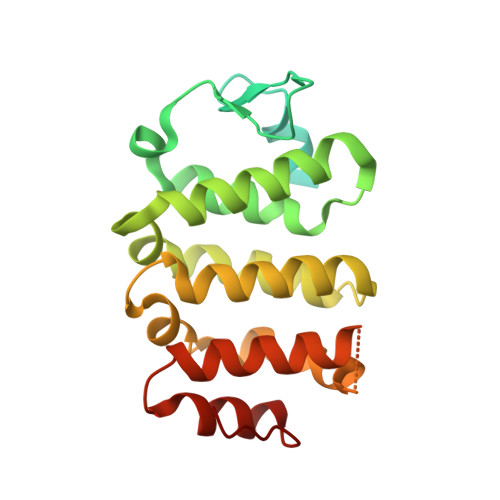 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q96M98 (Homo sapiens) Explore Q96M98 Go to UniProtKB: Q96M98 | |||||
PHAROS: Q96M98 GTEx: ENSG00000112530 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96M98 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Meiosis expressed gene 1 protein homolog | 93 | Homo sapiens | Mutation(s): 0 Gene Names: MEIG1 | 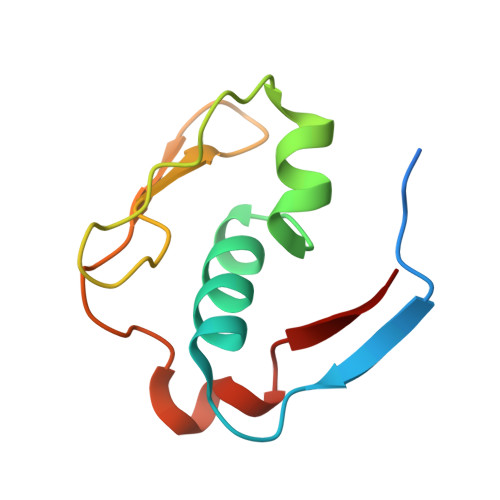 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q5JSS6 (Homo sapiens) Explore Q5JSS6 Go to UniProtKB: Q5JSS6 | |||||
PHAROS: Q5JSS6 GTEx: ENSG00000197889 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5JSS6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PG4 Query on PG4 | D [auth A] | TETRAETHYLENE GLYCOL C8 H18 O5 UWHCKJMYHZGTIT-UHFFFAOYSA-N |  | ||
| PEG Query on PEG | C [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| PO4 Query on PO4 | E [auth B] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 66.854 | α = 90 |
| b = 66.854 | β = 90 |
| c = 158.92 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHENIX | phasing |
Currently 6UCC does not have a validation slider image.
| Funding Organization | Location | Grant Number |
|---|---|---|
| Canadian Institutes of Health Research (CIHR) | Canada | 950-229792 X-239354 |