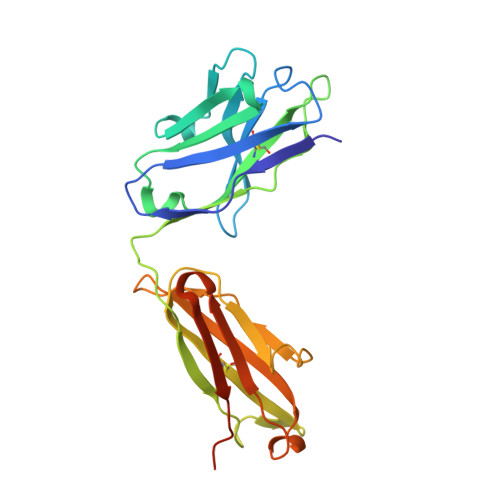
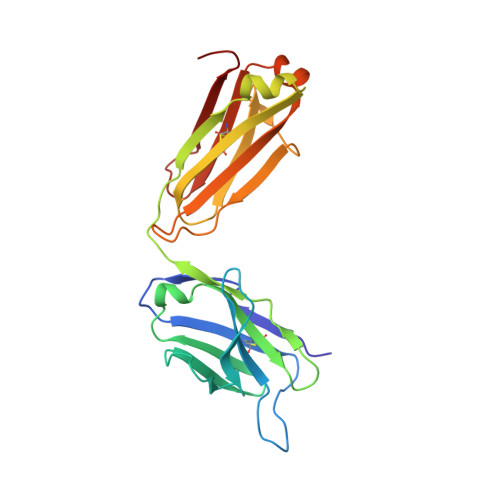
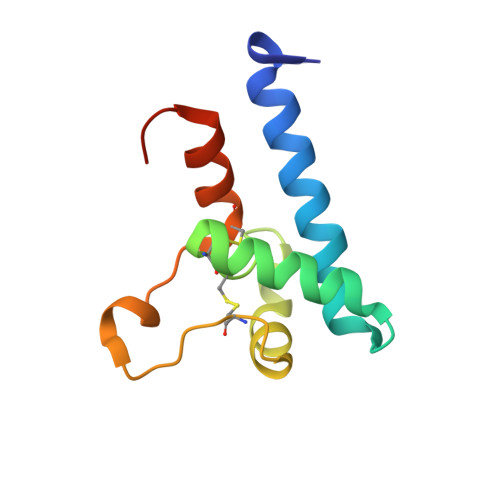
A dynamic interaction between CD19 and the tetraspanin CD81 controls B cell co-receptor trafficking.
Susa, K.J., Seegar, T.C., Blacklow, S.C., Kruse, A.C.(2020) Elife 9
- PubMed: 32338599
- DOI: https://doi.org/10.7554/eLife.52337
- Primary Citation of Related Structures:
6U9S - PubMed Abstract:
CD81 and its binding partner CD19 are core subunits of the B cell co-receptor complex. While CD19 belongs to the extensively studied Ig superfamily, CD81 belongs to a poorly understood family of four-pass transmembrane proteins called tetraspanins. Tetraspanins play important physiological roles by controlling protein trafficking and other processes. Here, we show that CD81 relies on its ectodomain to traffic CD19 to the cell surface. Moreover, the anti-CD81 antibody 5A6, which binds selectively to activated B cells, recognizes a conformational epitope on CD81 that is masked when CD81 is bound to CD19. Mutations of CD81 in this interface suppress its CD19 export activity. These data indicate that the CD81 - CD19 interaction is dynamically regulated upon B cell activation and this dynamism can be exploited to regulate B cell function. These results are not only valuable for understanding B cell biology, but also have important implications for understanding tetraspanin function generally.
- Department of Biological Chemistry and Molecular Pharmacology, Blavatnik Institute, Harvard Medical School, Boston, United States.
Organizational Affiliation: