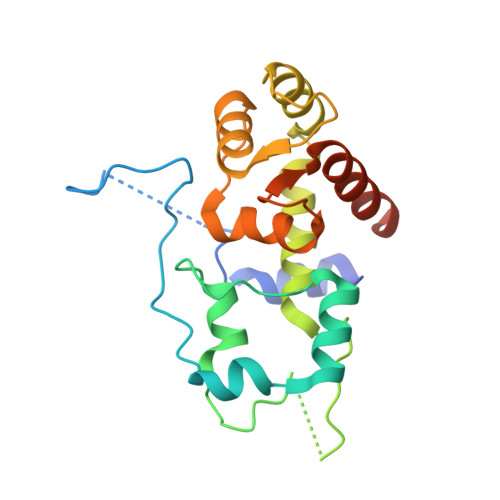
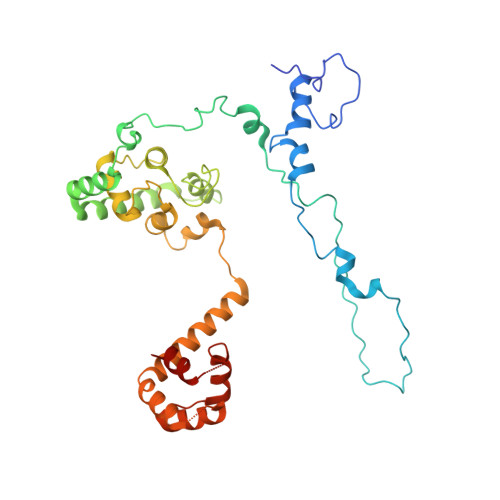
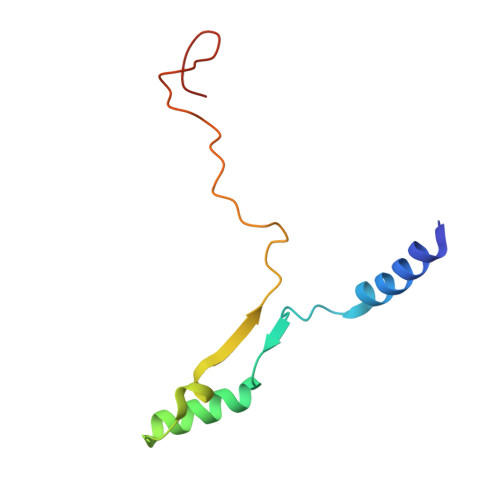
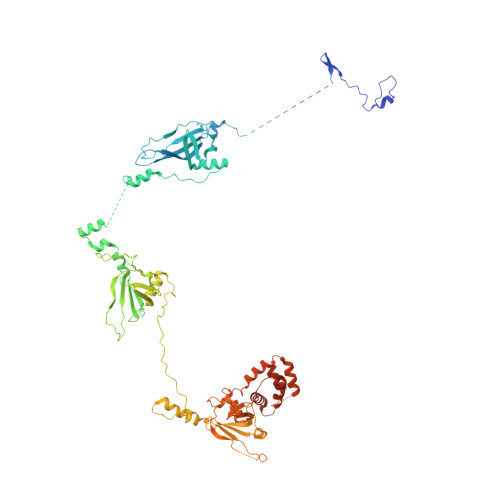
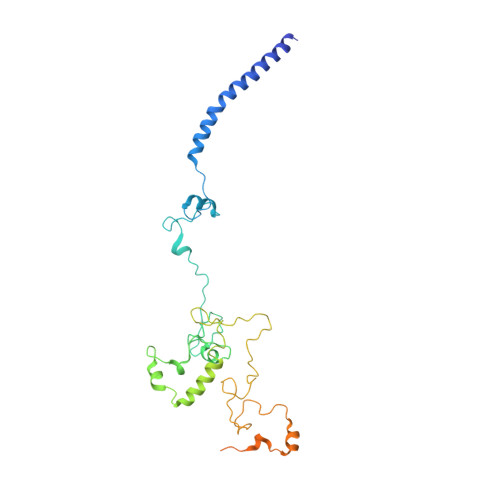
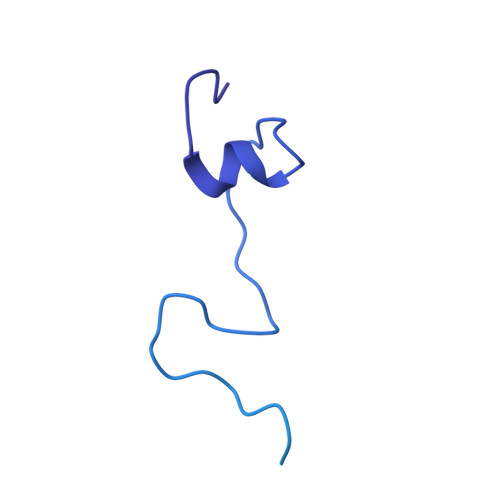
GTP Query on GTP Download Ideal Coordinates CCD File AAA [auth R2]
AAA [auth R2],
GUANOSINE-5'-TRIPHOSPHATE 10 H16 N5 O14 P3 GDP Query on GDP Download Ideal Coordinates CCD File ABA [auth T0]
ABA [auth T0],
GUANOSINE-5'-DIPHOSPHATE 10 H15 N5 O11 P2 MG Query on MG Download Ideal Coordinates CCD File ACA [auth U9]
ACA [auth U9],
MAGNESIUM ION