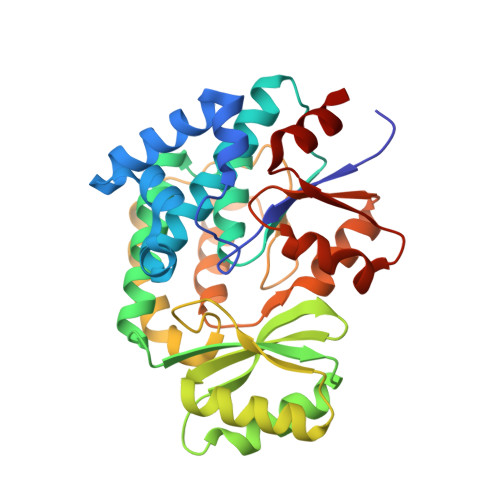
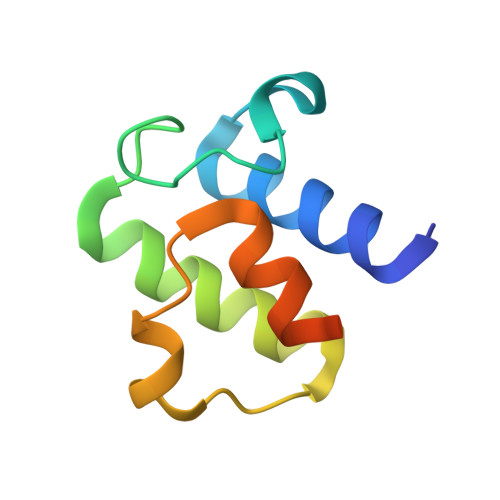
Interfacial plasticity facilitates high reaction rate of E. coli FAS malonyl-CoA:ACP transacylase, FabD.
Misson, L.E., Mindrebo, J.T., Davis, T.D., Patel, A., McCammon, J.A., Noel, J.P., Burkart, M.D.(2020) Proc Natl Acad Sci U S A 117: 24224-24233
- PubMed: 32929027
- DOI: https://doi.org/10.1073/pnas.2009805117
- Primary Citation of Related Structures:
6U0J - PubMed Abstract:
Fatty acid synthases (FASs) and polyketide synthases (PKSs) iteratively elongate and often reduce two-carbon ketide units in de novo fatty acid and polyketide biosynthesis. Cycles of chain extensions in FAS and PKS are initiated by an acyltransferase (AT), which loads monomer units onto acyl carrier proteins (ACPs), small, flexible proteins that shuttle covalently linked intermediates between catalytic partners. Formation of productive ACP-AT interactions is required for catalysis and specificity within primary and secondary FAS and PKS pathways. Here, we use the Escherichia coli FAS AT, FabD, and its cognate ACP, AcpP, to interrogate type II FAS ACP-AT interactions. We utilize a covalent crosslinking probe to trap transient interactions between AcpP and FabD to elucidate the X-ray crystal structure of a type II ACP-AT complex. Our structural data are supported using a combination of mutational, crosslinking, and kinetic analyses, and long-timescale molecular dynamics (MD) simulations. Together, these complementary approaches reveal key catalytic features of FAS ACP-AT interactions. These mechanistic inferences suggest that AcpP adopts multiple, productive conformations at the AT binding interface, allowing the complex to sustain high transacylation rates. Furthermore, MD simulations support rigid body subdomain motions within the FabD structure that may play a key role in AT activity and substrate selectivity.
- Department of Chemistry and Biochemistry, University of California San Diego, La Jolla, CA 92093-0358.
Organizational Affiliation: