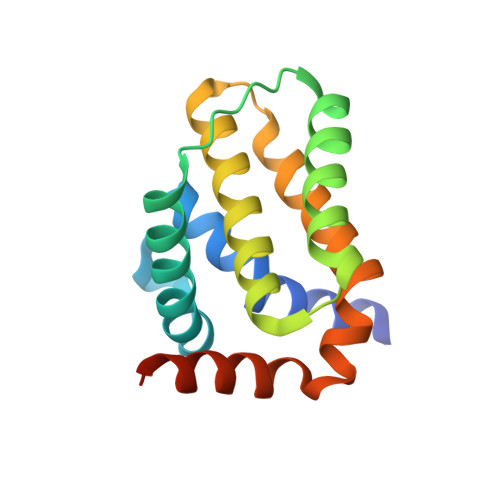
Crystal Structure of African Swine Fever Virus A179L with the Autophagy Regulator Beclin.
Banjara, S., Shimmon, G.L., Dixon, L.K., Netherton, C.L., Hinds, M.G., Kvansakul, M.(2019) Viruses 11
- PubMed: 31461953
- DOI: https://doi.org/10.3390/v11090789
- Primary Citation of Related Structures:
6TZC - PubMed Abstract:
Subversion of programmed cell death-based host defence systems is a prominent feature of infections by large DNA viruses. African swine fever virus (ASFV) is a large DNA virus and sole member of the Asfarviridae family that harbours the B-cell lymphoma 2 or Bcl-2 homolog A179L. A179L has been shown to bind to a range of cell death-inducing host proteins, including pro-apoptotic Bcl-2 proteins as well as the autophagy regulator Beclin. Here we report the crystal structure of A179L bound to the Beclin BH3 motif. A179L engages Beclin using the same canonical ligand-binding groove that is utilized to bind to pro-apoptotic Bcl-2 proteins. The mode of binding of Beclin to A179L mirrors that of Beclin binding to human Bcl-2 and Bcl-x L as well as murine γ-herpesvirus 68. The introduction of bulky hydrophobic residues into the A179L ligand-binding groove via site-directed mutagenesis ablates binding of Beclin to A179L, leading to a loss of the ability of A179L to modulate autophagosome formation in Vero cells during starvation. Our findings provide a mechanistic understanding for the potent autophagy inhibitory activity of A179L and serve as a platform for more detailed investigations into the role of autophagy during ASFV infection.
- Department of Biochemistry & Genetics, La Trobe Institute for Molecular Science, La Trobe University, Melbourne, Victoria 3086, Australia.
Organizational Affiliation: