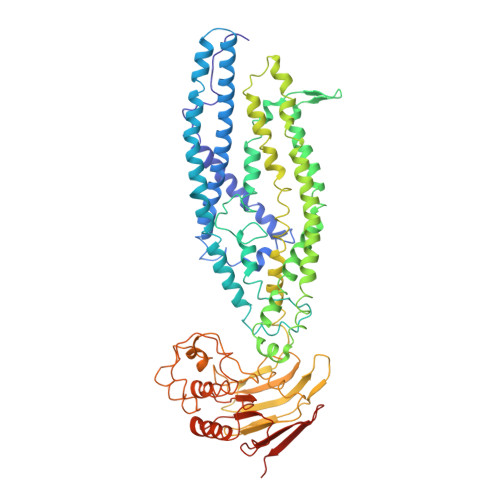
A mycobacterial ABC transporter mediates the uptake of hydrophilic compounds.
Rempel, S., Gati, C., Nijland, M., Thangaratnarajah, C., Karyolaimos, A., de Gier, J.W., Guskov, A., Slotboom, D.J.(2020) Nature 580: 409-412
- PubMed: 32296172
- DOI: https://doi.org/10.1038/s41586-020-2072-8
- Primary Citation of Related Structures:
6TQE, 6TQF - PubMed Abstract:
Mycobacterium tuberculosis (Mtb) is an obligate human pathogen and the causative agent of tuberculosis 1-3 . Although Mtb can synthesize vitamin B 12 (cobalamin) de novo, uptake of cobalamin has been linked to pathogenesis of tuberculosis 2 . Mtb does not encode any characterized cobalamin transporter 4-6 ; however, the gene rv1819c was found to be essential for uptake of cobalamin 1 . This result is difficult to reconcile with the original annotation of Rv1819c as a protein implicated in the transport of antimicrobial peptides such as bleomycin 7 . In addition, uptake of cobalamin seems inconsistent with the amino acid sequence, which suggests that Rv1819c has a bacterial ATP-binding cassette (ABC)-exporter fold 1 . Here, we present structures of Rv1819c, which reveal that the protein indeed contains the ABC-exporter fold, as well as a large water-filled cavity of about 7,700 Å 3 , which enables the protein to transport the unrelated hydrophilic compounds bleomycin and cobalamin. On the basis of these structures, we propose that Rv1819c is a multi-solute transporter for hydrophilic molecules, analogous to the multidrug exporters of the ABC transporter family, which pump out structurally diverse hydrophobic compounds from cells 8-11 .
- Groningen Biomolecular Sciences and Biotechnology Institute (GBB), University of Groningen, Groningen, The Netherlands.
Organizational Affiliation: