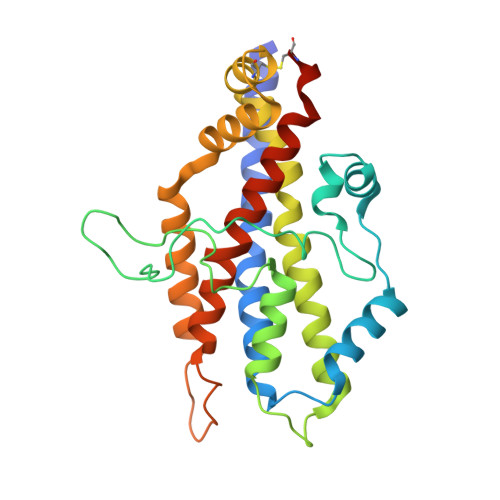
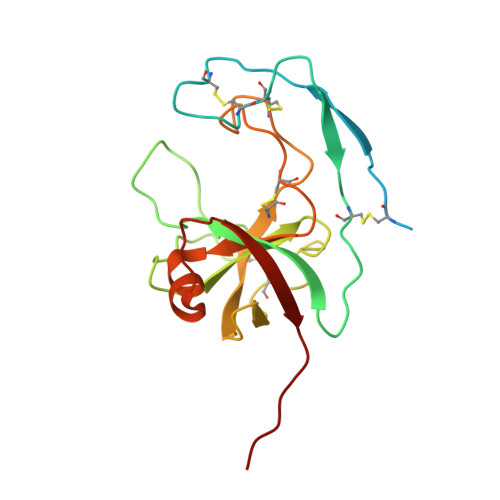
The two paralogous kiwellin proteins KWL1 and KWL1-b from maize are structurally related and have overlapping functions in plant defense.
Altegoer, F., Weiland, P., Giammarinaro, P.I., Freibert, S.A., Binnebesel, L., Han, X., Lepak, A., Kahmann, R., Lechner, M., Bange, G.(2020) J Biological Chem 295: 7816-7825
- PubMed: 32350112
- DOI: https://doi.org/10.1074/jbc.RA119.012207
- Primary Citation of Related Structures:
6TI2 - PubMed Abstract:
Many plant-pathogenic bacteria and fungi deploy effector proteins that down-regulate plant defense responses and reprogram plant metabolism for colonization and survival in planta Kiwellin (KWL) proteins are a widespread family of plant-defense proteins that target these microbial effectors. The KWL1 protein from maize (corn, Zea mays ) specifically inhibits the enzymatic activity of the secreted chorismate mutase Cmu1, a virulence-promoting effector of the smut fungus Ustilago maydis. In addition to KWL1, 19 additional KWL paralogs have been identified in maize. Here, we investigated the structure and mechanism of the closest KWL1 homolog, KWL1-b (ZEAMA_GRMZM2G305329). We solved the Cmu1-KWL1-b complex to 2.75 Å resolution, revealing a highly symmetric Cmu1-KWL1-b heterotetramer in which each KWL1-b monomer interacts with a monomer of the Cmu1 homodimer. The structure also revealed that the overall architecture of the heterotetramer is highly similar to that of the previously reported Cmu1-KWL1 complex. We found that upon U. maydis infection of Z. mays , KWL1-b is expressed at significantly lower levels than KWL1 and exhibits differential tissue-specific expression patterns. We also show that KWL1-b inhibits Cmu1 activity similarly to KWL1. We conclude that KWL1 and KWL1-b are part of a redundant defense system that tissue-specifically targets Cmu1. This notion was supported by the observation that both KWL proteins are carbohydrate-binding proteins with distinct and likely tissue-related specificities. Moreover, binding by Cmu1 modulated the carbohydrate-binding properties of both KWLs. These findings indicate that KWL proteins are part of a spatiotemporally coordinated, plant-wide defense response comprising proteins with overlapping activities.
- SYNMIKRO Research Center and Department of Chemistry, Philipps-University Marburg, Marburg, Germany florian.altegoer@synmikro.uni-marburg.de gert.bange@synmikro.uni-marburg.de.
Organizational Affiliation: