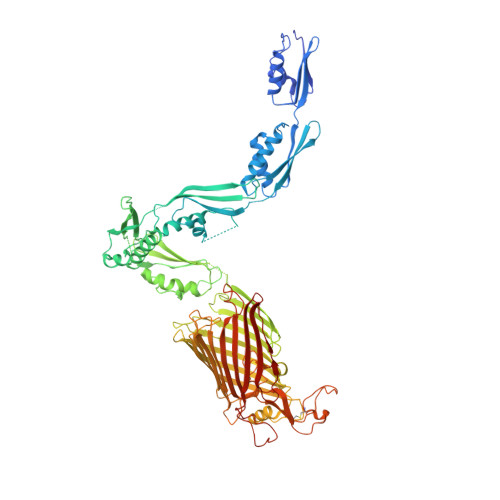
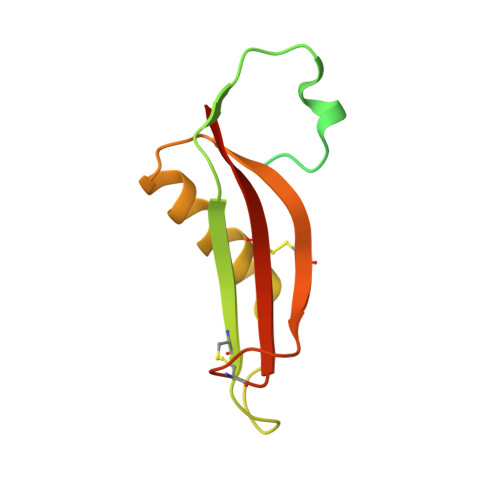
Structural insight into the formation of lipoprotein-beta-barrel complexes.
Rodriguez-Alonso, R., Letoquart, J., Nguyen, V.S., Louis, G., Calabrese, A.N., Iorga, B.I., Radford, S.E., Cho, S.H., Remaut, H., Collet, J.F.(2020) Nat Chem Biol 16: 1019-1025
- PubMed: 32572278
- DOI: https://doi.org/10.1038/s41589-020-0575-0
- Primary Citation of Related Structures:
6T1W - PubMed Abstract:
The β-barrel assembly machinery (BAM) inserts outer membrane β-barrel proteins (OMPs) in the outer membrane of Gram-negative bacteria. In Enterobacteriacea, BAM also mediates export of the stress sensor lipoprotein RcsF to the cell surface by assembling RcsF-OMP complexes. Here, we report the crystal structure of the key BAM component BamA in complex with RcsF. BamA adopts an inward-open conformation, with the lateral gate to the membrane closed. RcsF is lodged deep within the lumen of the BamA barrel, binding regions proposed to undergo outward and lateral opening during OMP insertion. On the basis of our structural and biochemical data, we propose a push-and-pull model for RcsF export following conformational cycling of BamA, and provide a mechanistic explanation for how RcsF uses its interaction with BamA to detect envelope stress. Our data also suggest that the flux of incoming OMP substrates is involved in the control of BAM activity.
- Walloon Excellence in Life Sciences and Biotechnology, Brussels, Belgium.
Organizational Affiliation: