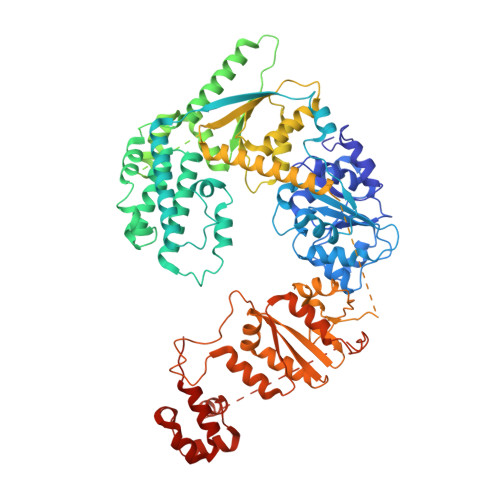
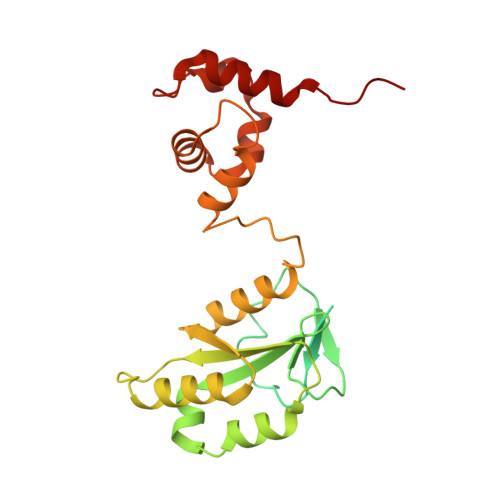
Cryo-EM structures of the XPF-ERCC1 endonuclease reveal how DNA-junction engagement disrupts an auto-inhibited conformation.
Jones, M., Beuron, F., Borg, A., Nans, A., Earl, C.P., Briggs, D.C., Snijders, A.P., Bowles, M., Morris, E.P., Linch, M., McDonald, N.Q.(2020) Nat Commun 11: 1120-1120
- PubMed: 32111838
- DOI: https://doi.org/10.1038/s41467-020-14856-2
- Primary Citation of Related Structures:
6SXA, 6SXB - PubMed Abstract:
The structure-specific endonuclease XPF-ERCC1 participates in multiple DNA damage repair pathways including nucleotide excision repair (NER) and inter-strand crosslink repair (ICLR). How XPF-ERCC1 is catalytically activated by DNA junction substrates is not currently understood. Here we report cryo-electron microscopy structures of both DNA-free and DNA-bound human XPF-ERCC1. DNA-free XPF-ERCC1 adopts an auto-inhibited conformation in which the XPF helical domain masks the ERCC1 (HhH) 2 domain and restricts access to the XPF catalytic site. DNA junction engagement releases the ERCC1 (HhH) 2 domain to couple with the XPF-ERCC1 nuclease/nuclease-like domains. Structure-function data indicate xeroderma pigmentosum patient mutations frequently compromise the structural integrity of XPF-ERCC1. Fanconi anaemia patient mutations in XPF often display substantial in-vitro activity but are resistant to activation by ICLR recruitment factor SLX4. Our data provide insights into XPF-ERCC1 architecture and catalytic activation.
- Signalling and Structural Biology Laboratory, Francis Crick Institute, NW1 1AT, London, UK.
Organizational Affiliation: