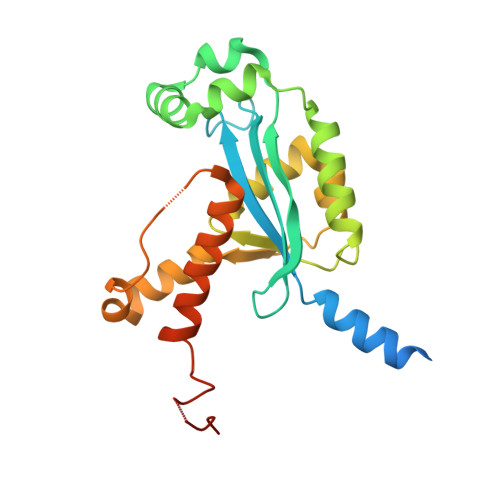
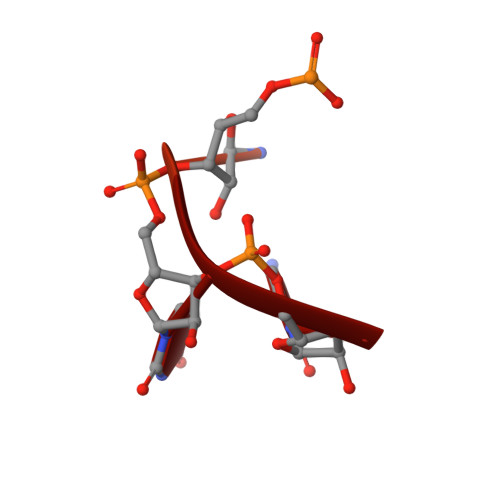
Human REXO2 controls short mitochondrial RNAs generated by mtRNA processing and decay machinery to prevent accumulation of double-stranded RNA.
Szewczyk, M., Malik, D., Borowski, L.S., Czarnomska, S.D., Kotrys, A.V., Klosowska-Kosicka, K., Nowotny, M., Szczesny, R.J.(2020) Nucleic Acids Res 48: 5572-5590
- PubMed: 32365187
- DOI: https://doi.org/10.1093/nar/gkaa302
- Primary Citation of Related Structures:
6STY - PubMed Abstract:
RNA decay is a key element of mitochondrial RNA metabolism. To date, the only well-documented machinery that plays a role in mtRNA decay in humans is the complex of polynucleotide phosphorylase (PNPase) and SUV3 helicase, forming the degradosome. REXO2, a homolog of prokaryotic oligoribonucleases present in humans both in mitochondria and the cytoplasm, was earlier shown to be crucial for maintaining mitochondrial homeostasis, but its function in mitochondria has not been fully elucidated. In the present study, we created a cellular model that enables the clear dissection of mitochondrial and non-mitochondrial functions of human REXO2. We identified a novel mitochondrial short RNA, referred to as ncH2, that massively accumulated upon REXO2 silencing. ncH2 degradation occurred independently of the mitochondrial degradosome, strongly supporting the hypothesis that ncH2 is a primary substrate of REXO2. We also investigated the global impact of REXO2 depletion on mtRNA, revealing the importance of the protein for maintaining low steady-state levels of mitochondrial antisense transcripts and double-stranded RNA. Our detailed biochemical and structural studies provide evidence of sequence specificity of the REXO2 oligoribonuclease. We postulate that REXO2 plays dual roles in human mitochondria, 'scavenging' nanoRNAs that are produced by the degradosome and clearing short RNAs that are generated by RNA processing.
- Institute of Biochemistry and Biophysics Polish Academy of Sciences, Warsaw 02-106, Poland.
Organizational Affiliation: