The specific biochemistry of human axilla odour formation viewed in an evolutionary context.
Natsch, A., Emter, R.(2020) Philos Trans R Soc Lond B Biol Sci 375: 20190269-20190269
- PubMed: 32306870
- DOI: https://doi.org/10.1098/rstb.2019.0269
- Primary Citation of Related Structures:
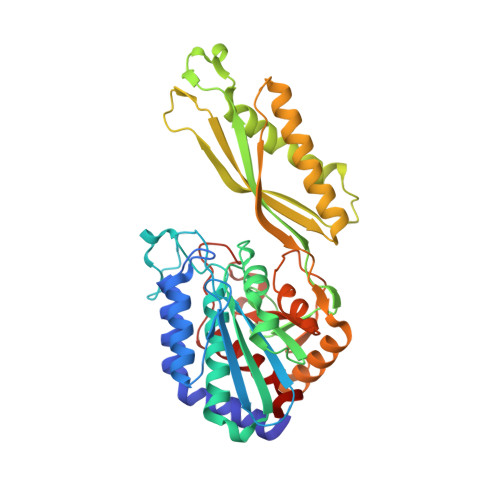
6SLF - PubMed Abstract:
Human body odour is dominated by the scent of specific odourants emanating from specialized glands in the axillary region. These specific odourants are produced by an intricate interplay between biochemical pathways in the host and odour-releasing enzymes present in commensal microorganisms of the axillary microbiome. Key biochemical steps for the release of highly odouriferous carboxylic acids and sulfur compounds have been elucidated over the past 15 years. Based on the profound molecular understanding and specific analytical methods developed, evolutionary questions could be asked for the first time with small population studies: (i) a genetic basis for body odour could be shown with a twin study, (ii) no effect of genes in the human leukocyte antigen complex on the pattern of odourant carboxylic acid was found, and (iii) loss of odour precursor secretion by a mutation in the ABCC11 gene could explain why a large fraction of the population in the Far East lack body odour formation. This review summarizes what is currently known at the molecular level on the biochemistry of the formation of key odourants in the human axilla. At the same time, we present for the first time the crystal structure of the N α -acyl-aminoacylase, a key human odour-releasing enzyme, thus describing at the molecular level how bacteria on the skin surface have adapted their enzyme to the specific substrates secreted by the human host. This article is part of the Theo Murphy meeting issue 'Olfactory communication in humans'.
- Givaudan Schweiz AG, Kemptpark 50, CH-8310 Kemptthal, Switzerland.
Organizational Affiliation: