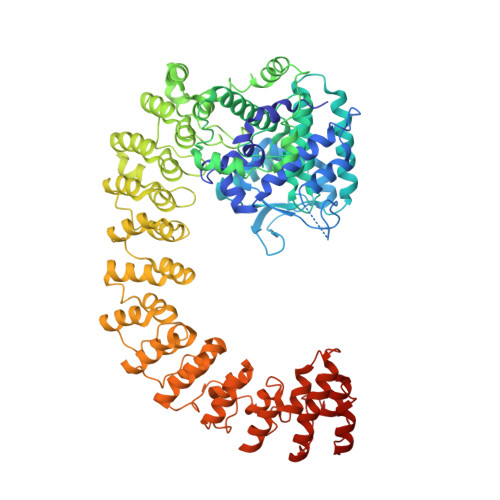
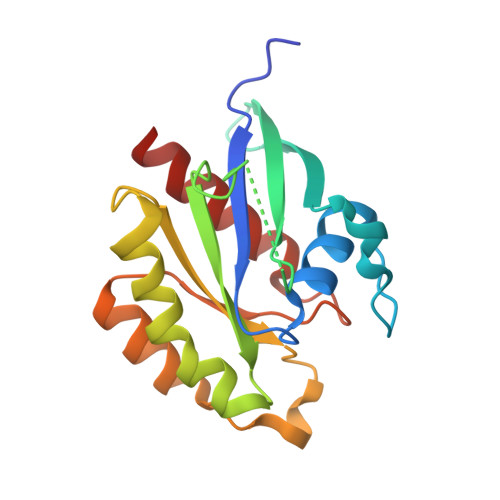
Legionellaeffector AnkX displaces the switch II region for Rab1b phosphocholination.
Ernst, S., Ecker, F., Kaspers, M.S., Ochtrop, P., Hedberg, C., Groll, M., Itzen, A.(2020) Sci Adv 6: eaaz8041-eaaz8041
- PubMed: 32440549
- DOI: https://doi.org/10.1126/sciadv.aaz8041
- Primary Citation of Related Structures:
6SKU - PubMed Abstract:
The causative agent of Legionnaires disease, Legionella pneumophila , translocates the phosphocholine transferase AnkX during infection and thereby posttranslationally modifies the small guanosine triphosphatase (GTPase) Rab1 with a phosphocholine moiety at S76 using cytidine diphosphate (CDP)-choline as a cosubstrate. The molecular basis for Rab1 binding and enzymatic modification have remained elusive because of lack of structural information of the low-affinity complex with AnkX. We combined thiol-reactive CDP-choline derivatives with recombinantly introduced cysteines in the AnkX active site to covalently capture the heterocomplex. The resulting crystal structure revealed that AnkX induces displacement of important regulatory elements of Rab1 by placing a β sheet into a conserved hydrophobic pocket, thereby permitting phosphocholine transfer to the active and inactive states of the GTPase. Together, the combination of chemical biology and structural analysis reveals the enzymatic mechanism of AnkX and the family of filamentation induced by cyclic adenosine monophosphate (FIC) proteins.
- Department of Biochemistry and Signal Transduction, University Medical Centre Hamburg-Eppendorf (UKE), Martinistr. 52, 20246 Hamburg, Germany.
Organizational Affiliation: