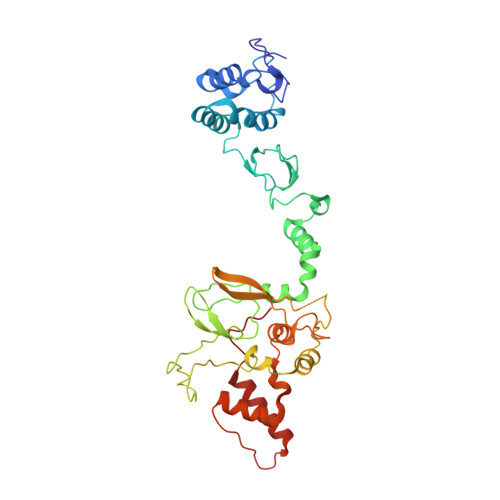
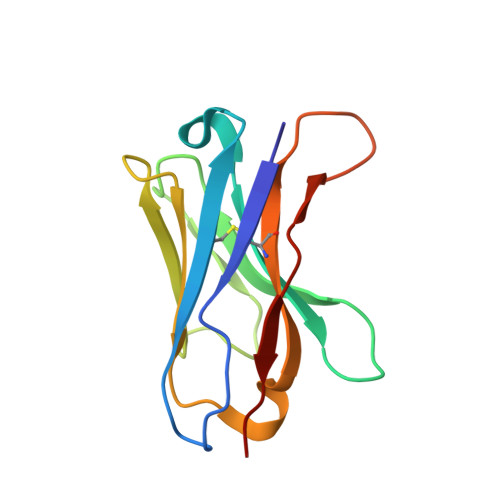
Single-Domain Antibodies as Crystallization Chaperones to Enable Structure-Based Inhibitor Development for RBR E3 Ubiquitin Ligases.
Tsai, Y.I., Johansson, H., Dixon, D., Martin, S., Chung, C.W., Clarkson, J., House, D., Rittinger, K.(2020) Cell Chem Biol 27: 83
- PubMed: 31813847
- DOI: https://doi.org/10.1016/j.chembiol.2019.11.007
- Primary Citation of Related Structures:
6SC5, 6SC6, 6SC7, 6SC8, 6SC9, 6T2J - PubMed Abstract:
Protein ubiquitination plays a key role in the regulation of cellular processes, and misregulation of the ubiquitin system is linked to many diseases. So far, development of tool compounds that target enzymes of the ubiquitin system has been slow and only a few specific inhibitors are available. Here, we report the selection of single-domain antibodies (single-dAbs) based on a human scaffold that recognize the catalytic domain of HOIP, a subunit of the multi-component E3 LUBAC and member of the RBR family of E3 ligases. Some of these dAbs affect ligase activity and provide mechanistic insight into the ubiquitin transfer mechanism of different E2-conjugating enzymes. Furthermore, we show that the co-crystal structure of a HOIP RBR/dAb complex serves as a robust platform for soaking of ligands that target the active site cysteine of HOIP, thereby providing easy access to structure-based ligand design for this important class of E3 ligases.
- Molecular Structure of Cell Signalling Laboratory, The Francis Crick Institute, 1 Midland Road, London NW1 1AT, UK.
Organizational Affiliation: