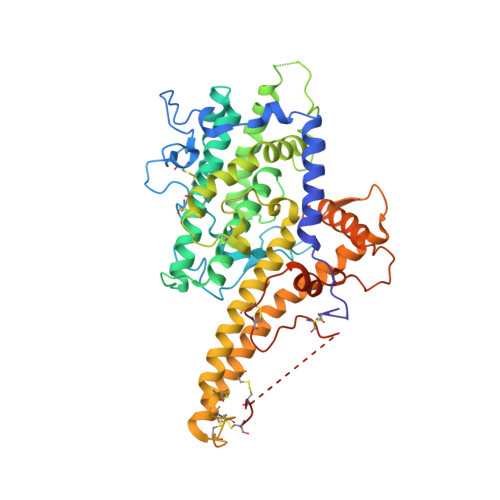
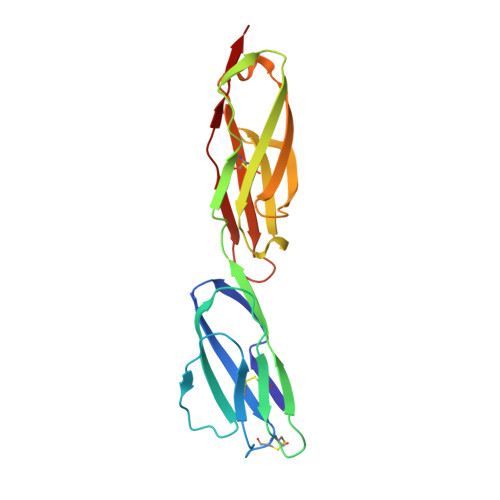
Structural insights into diverse modes of ICAM-1 binding byPlasmodium falciparum-infected erythrocytes.
Lennartz, F., Smith, C., Craig, A.G., Higgins, M.K.(2019) Proc Natl Acad Sci U S A 116: 20124-20134
- PubMed: 31527263
- DOI: https://doi.org/10.1073/pnas.1911900116
- Primary Citation of Related Structures:
6S8T, 6S8U - PubMed Abstract:
A major determinant of pathogenicity in malaria caused by Plasmodium falciparum is the adhesion of parasite-infected erythrocytes to the vasculature or tissues of infected individuals. This occludes blood flow, leads to inflammation, and increases parasitemia by reducing spleen-mediated clearance of the parasite. This adhesion is mediated by PfEMP1, a multivariant family of around 60 proteins per parasite genome which interact with specific host receptors. One of the most common of these receptors is intracellular adhesion molecule-1 (ICAM-1), which is bound by 2 distinct groups of PfEMP1, A-type and B or C (BC)-type. Here, we present the structure of a domain from a B-type PfEMP1 bound to ICAM-1, revealing a complex binding site. Comparison with the existing structure of an A-type PfEMP1 bound to ICAM-1 shows that the 2 complexes share a globally similar architecture. However, while the A-type PfEMP1 bind ICAM-1 through a highly conserved binding surface, the BC-type PfEMP1 use a binding site that is more diverse in sequence, similar to how PfEMP1 interact with other human receptors. We also show that A- and BC-type PfEMP1 present ICAM-1 at different angles, perhaps influencing the ability of neighboring PfEMP1 domains to bind additional receptors. This illustrates the deep diversity of the PfEMP1 and demonstrates how variations in a single domain architecture can modulate binding to a specific ligand to control function and facilitate immune evasion.
- Department of Biochemistry, University of Oxford, OX1 3QU Oxford, United Kingdom.
Organizational Affiliation: