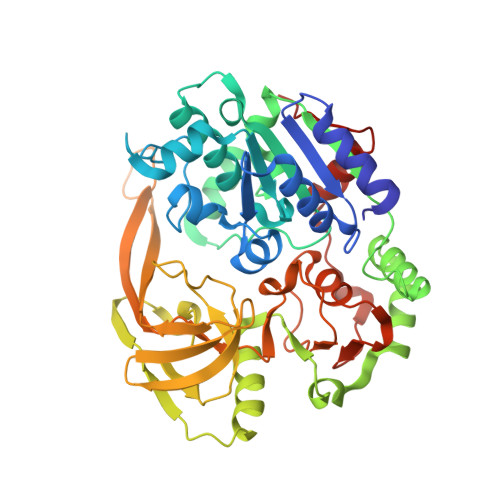
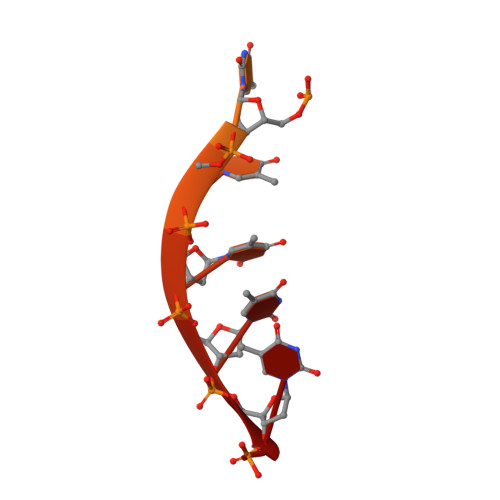
Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Dai, Y.X., Chen, W.F., Liu, N.N., Teng, F.Y., Guo, H.L., Hou, X.M., Dou, S.X., Rety, S., Xi, X.G.(2021) Nucleic Acids Res 49: 4129-4143
- PubMed: 33784404
- DOI: https://doi.org/10.1093/nar/gkab188
- Primary Citation of Related Structures:
6S3E, 6S3H, 6S3I, 6S3M, 6S3N, 6S3O, 6S3P, 6XZT, 7ADA, 7BIL - PubMed Abstract:
Pif1 is an SF1B helicase that is evolutionarily conserved from bacteria to humans and plays multiple roles in maintaining genome stability in both nucleus and mitochondria. Though highly conserved, Pif1 family harbors a large mechanistic diversity. Here, we report crystal structures of Thermus oshimai Pif1 (ToPif1) alone and complexed with partial duplex or single-stranded DNA. In the apo state and in complex with a partial duplex DNA, ToPif1 is monomeric with its domain 2B/loop3 adopting a closed and an open conformation, respectively. When complexed with a single-stranded DNA, ToPif1 forms a stable dimer with domain 2B/loop3 shifting to a more open conformation. Single-molecule and biochemical assays show that domain 2B/loop3 switches repetitively between the closed and open conformations when a ToPif1 monomer unwinds DNA and, in contrast with other typical dimeric SF1A helicases, dimerization has an inhibitory effect on its helicase activity. This mechanism is not general for all Pif1 helicases but illustrates the diversity of regulation mechanisms among different helicases. It also raises the possibility that although dimerization results in activation for SF1A helicases, it may lead to inhibition for some of the other uncharacterized SF1B helicases, an interesting subject warranting further studies.
- State Key Laboratory of Crop Stress Biology for Arid Areas and College of Life Sciences, Northwest A&F University, Yangling, Shaanxi 712100, China.
Organizational Affiliation: