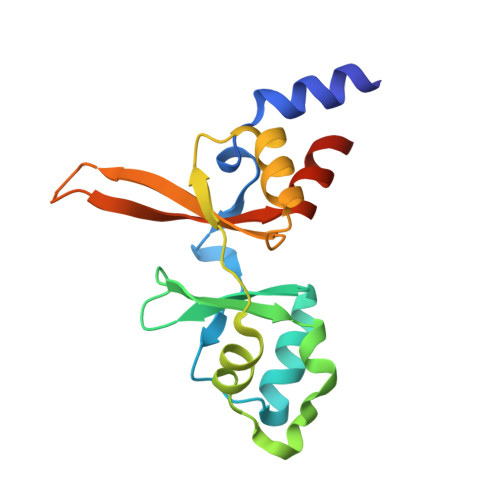
Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Gimenez-Mascarell, P., Oyenarte, I., Gonzalez-Recio, I., Fernandez-Rodriguez, C., Corral-Rodriguez, M.A., Campos-Zarraga, I., Simon, J., Kostantin, E., Hardy, S., Diaz Quintana, A., Zubillaga Lizeaga, M., Merino, N., Diercks, T., Blanco, F.J., Diaz Moreno, I., Martinez-Chantar, M.L., Tremblay, M.L., Muller, D., Siliqi, D., Martinez-Cruz, L.A.(2019) Int J Mol Sci 20
- PubMed: 31842432
- DOI: https://doi.org/10.3390/ijms20246279
- Primary Citation of Related Structures:
6G52, 6RS2 - PubMed Abstract:
The four member family of "Cyclin and Cystathionine β-synthase (CBS) domain divalent metal cation transport mediators", CNNMs, are the least-studied mammalian magnesium transport mediators. CNNM4 is abundant in the brain and the intestinal tract, and its abnormal activity causes Jalili Syndrome. Recent findings show that suppression of CNNM4 in mice promotes malignant progression of intestinal polyps and is linked to infertility. The association of CNNM4 with phosphatases of the regenerating liver, PRLs, abrogates its Mg 2+ -efflux capacity, thus resulting in an increased intracellular Mg 2+ concentration that favors tumor growth. Here we present the crystal structures of the two independent intracellular domains of human CNNM4, i.e., the Bateman module and the cyclic nucleotide binding-like domain (cNMP). We also derive a model structure for the full intracellular region in the absence and presence of MgATP and the oncogenic interacting partner, PRL-1. We find that only the Bateman module interacts with ATP and Mg 2+ , at non-overlapping sites facilitating their positive cooperativity. Furthermore, both domains dimerize autonomously, where the cNMP domain dimer forms a rigid cleft to restrict the Mg 2+ induced sliding of the inserting CBS1 motives of the Bateman module, from a twisted to a flat disk shaped dimer.
- Liver Disease Laboratory, Center for Cooperative Research in Biosciences (CIC bioGUNE), Bizkaia Science and Technology Park Bld 801A, 48160 Derio, Spain.
Organizational Affiliation: