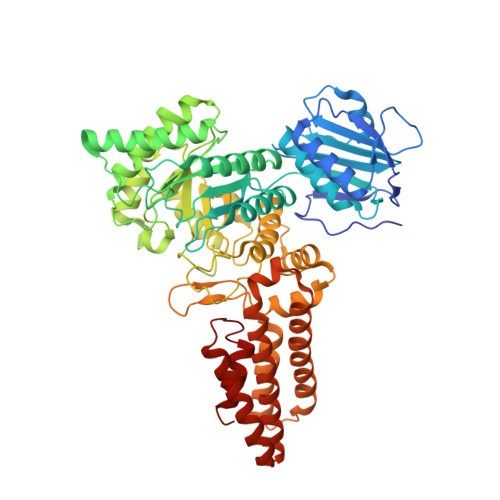
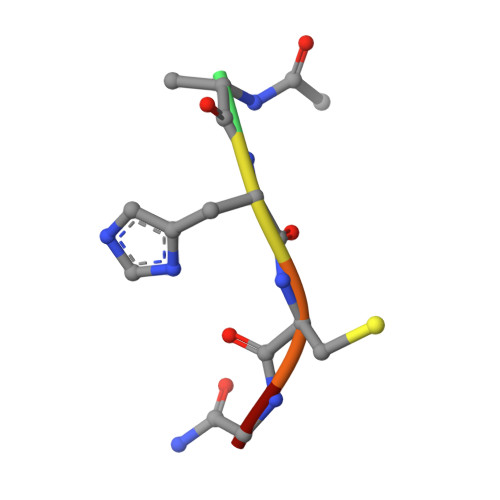
Genetic recoding to dissect the roles of site-specific protein O-GlcNAcylation.
Gorelik, A., Bartual, S.G., Borodkin, V.S., Varghese, J., Ferenbach, A.T., van Aalten, D.M.F.(2019) Nat Struct Mol Biol 26: 1071-1077
- PubMed: 31695185
- DOI: https://doi.org/10.1038/s41594-019-0325-8
- Primary Citation of Related Structures:
6RHE - PubMed Abstract:
Modification of specific Ser and Thr residues of nucleocytoplasmic proteins with O-GlcNAc, catalyzed by O-GlcNAc transferase (OGT), is an abundant posttranslational event essential for proper animal development and is dysregulated in various diseases. Due to the rapid concurrent removal by the single O-GlcNAcase (OGA), precise functional dissection of site-specific O-GlcNAc modification in vivo is currently not possible without affecting the entire O-GlcNAc proteome. Exploiting the fortuitous promiscuity of OGT, we show that S-GlcNAc is a hydrolytically stable and accurate structural mimic of O-GlcNAc that can be encoded in mammalian systems with CRISPR-Cas9 in an otherwise unperturbed O-GlcNAcome. Using this approach, we target an elusive Ser 405 O-GlcNAc site on OGA, showing that this site-specific modification affects OGA stability.
- Centre for Gene Regulation and Expression, School of Life Sciences, University of Dundee, Dundee, UK.
Organizational Affiliation: