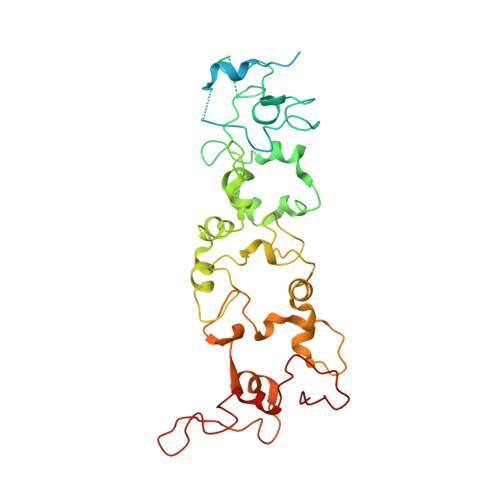
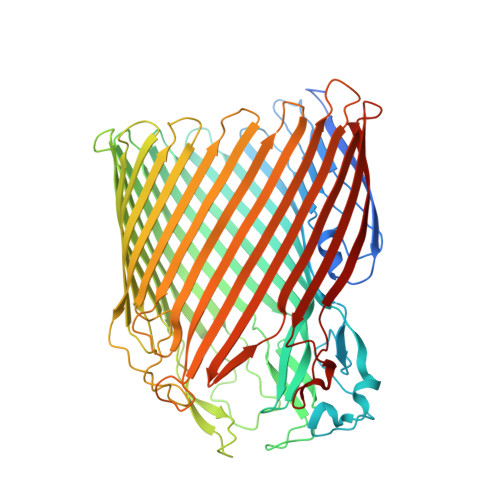
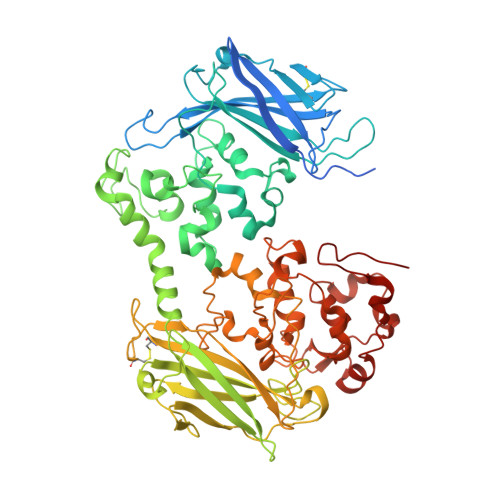
The Crystal Structure of a Biological Insulated Transmembrane Molecular Wire.
Edwards, M.J., White, G.F., Butt, J.N., Richardson, D.J., Clarke, T.A.(2020) Cell 181: 665-673.e10
- PubMed: 32289252
- DOI: https://doi.org/10.1016/j.cell.2020.03.032
- Primary Citation of Related Structures:
6QYC, 6R2Q - PubMed Abstract:
A growing number of bacteria are recognized to conduct electrons across their cell envelope, and yet molecular details of the mechanisms supporting this process remain unknown. Here, we report the atomic structure of an outer membrane spanning protein complex, MtrAB, that is representative of a protein family known to transport electrons between the interior and exterior environments of phylogenetically and metabolically diverse microorganisms. The structure is revealed as a naturally insulated biomolecular wire possessing a 10-heme cytochrome, MtrA, insulated from the membrane lipidic environment by embedding within a 26 strand β-barrel formed by MtrB. MtrAB forms an intimate connection with an extracellular 10-heme cytochrome, MtrC, which presents its hemes across a large surface area for electrical contact with extracellular redox partners, including transition metals and electrodes.
- School of Biological Sciences, University of East Anglia, Norwich NR4 7TJ, UK.
Organizational Affiliation: