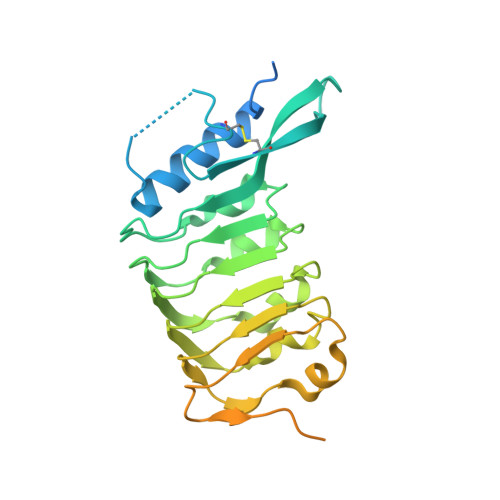
Crystal structure of the leucine-rich repeat ectodomain of the plant immune receptor kinase SOBIR1.
Hohmann, U., Hothorn, M.(2019) Acta Crystallogr D Struct Biol 75: 488-497
- PubMed: 31063151
- DOI: https://doi.org/10.1107/S2059798319005291
- Primary Citation of Related Structures:
6R1H - PubMed Abstract:
Plant-unique membrane receptor kinases with leucine-rich repeat (LRR) extracellular domains are key regulators of development and immune responses. Here, the 1.55 Å resolution crystal structure of the immune receptor kinase SOBIR1 from Arabidopsis is presented. The ectodomain structure reveals the presence of five LRRs sandwiched between noncanonical capping domains. The disulfide-bond-stabilized N-terminal cap harbours an unusual β-hairpin structure. The C-terminal cap features a highly positively charged linear motif which was found to be largely disordered in this structure. Size-exclusion chromatography and right-angle light-scattering experiments suggest that SOBIR1 is a monomer in solution. The protruding β-hairpin, a set of highly conserved basic residues at the inner surface of the SOBIR LRR domain and the presence of a genetic missense allele in LRR2 together suggest that the SOBIR1 ectodomain may mediate protein-protein interaction in plant immune signalling.
- Structural Plant Biology Laboratory, Department of Botany and Plant Biology, University of Geneva, 1211 Geneva, Switzerland.
Organizational Affiliation: