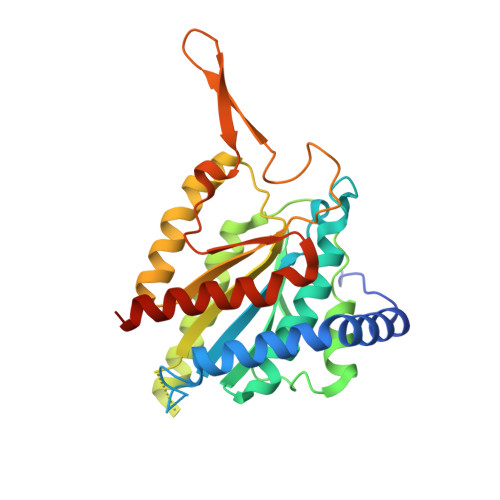
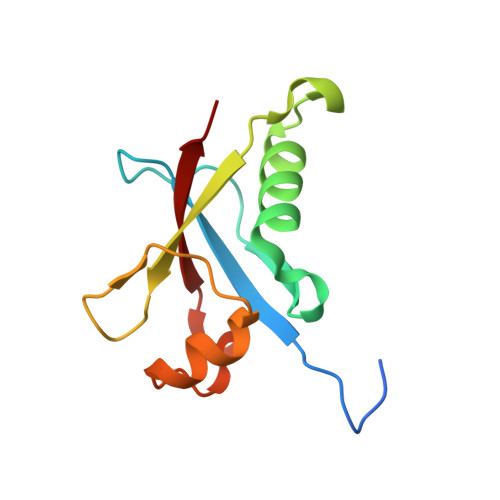
Crystal structures of Rea1-MIDAS bound to its ribosome assembly factor ligands resembling integrin-ligand-type complexes.
Ahmed, Y.L., Thoms, M., Mitterer, V., Sinning, I., Hurt, E.(2019) Nat Commun 10: 3050-3050
- PubMed: 31296859
- DOI: https://doi.org/10.1038/s41467-019-10922-6
- Primary Citation of Related Structures:
6QT8, 6QTA, 6QTB - PubMed Abstract:
The Rea1 AAA + -ATPase dislodges assembly factors from pre-60S ribosomes upon ATP hydrolysis, thereby driving ribosome biogenesis. Here, we present crystal structures of Rea1-MIDAS, the conserved domain at the tip of the flexible Rea1 tail, alone and in complex with its substrate ligands, the UBL domains of Rsa4 or Ytm1. These complexes have structural similarity to integrin α-subunit domains when bound to extracellular matrix ligands, which for integrin biology is a key determinant for force-bearing cell-cell adhesion. However, the presence of additional motifs equips Rea1-MIDAS for its tasks in ribosome maturation. One loop insert cofunctions as an NLS and to activate the mechanochemical Rea1 cycle, whereas an additional β-hairpin provides an anchor to hold the ligand UBL domains in place. Our data show the versatility of the MIDAS fold for mechanical force transmission in processes as varied as integrin-mediated cell adhesion and mechanochemical removal of assembly factors from pre-ribosomes.
- Heidelberg University Biochemistry Center, D-69120, Heidelberg, Germany.
Organizational Affiliation: