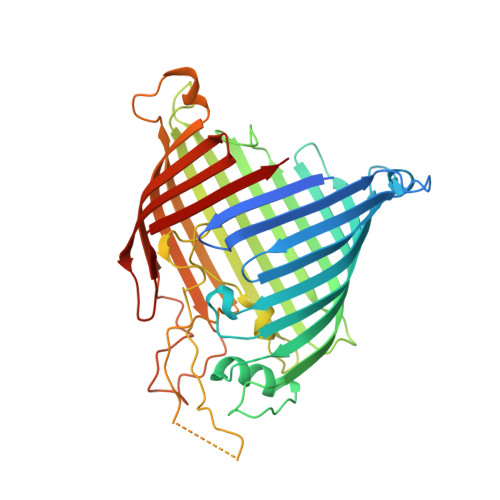
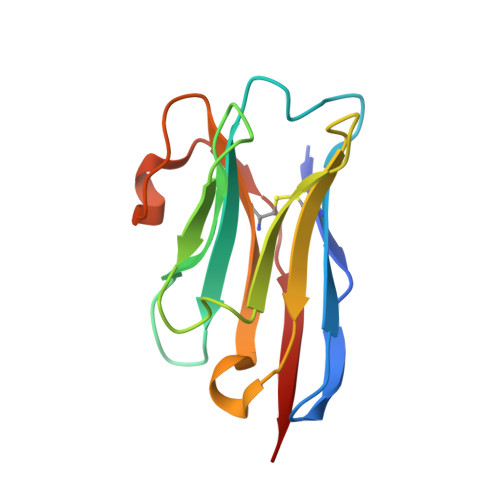
Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
Kaur, H., Hartmann, J.B., Jakob, R.P., Zahn, M., Zimmermann, I., Maier, T., Seeger, M.A., Hiller, S.(2019) J Biomol NMR 73: 375-384
- PubMed: 31073665
- DOI: https://doi.org/10.1007/s10858-019-00250-8
- Primary Citation of Related Structures:
6QGW, 6QGX, 6QGY - PubMed Abstract:
The insertase BamA is an essential protein of the bacterial outer membrane. Its 16-stranded transmembrane β-barrel contains a lateral gate as a key functional element. This gate is formed by the C-terminal half of the last β-strand. The BamA barrel was previously found to sample different conformations in aqueous solution, as well as different gate-open, gate-closed, and collapsed conformations in X-ray crystallography and cryo-electron microscopy structures. Here, we report the successful identification of conformation-selective nanobodies that stabilize BamA in specific conformations. While the initial candidate generation and selection protocol was based on established alpaca immunization and phage display selection procedures, the final selection of nanobodies was enhanced by a solution NMR-based screening step to shortlist the targets for crystallization. In this way, three crystal structures of BamA-nanobody complexes were efficiently obtained, showing two types of nanobodies that indeed stabilized BamA in two different conformations, i.e., with open and closed lateral gate, respectively. Then, by correlating the structural data with high resolution NMR spectra, we could for the first time assign the BamA conformational solution ensemble to defined structural states. The new nanobodies will be valuable tools towards understanding the client insertion mechanism of BamA and towards developing improved antibiotics.
- Biozentrum, University of Basel, 4056, Basel, Switzerland.
Organizational Affiliation: