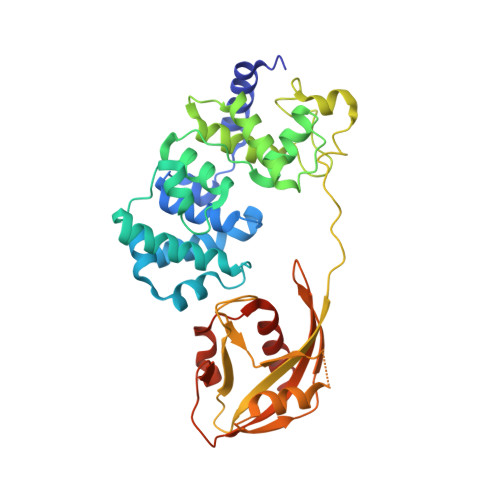
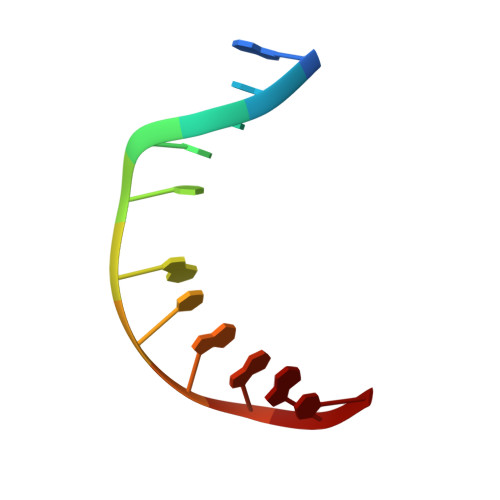
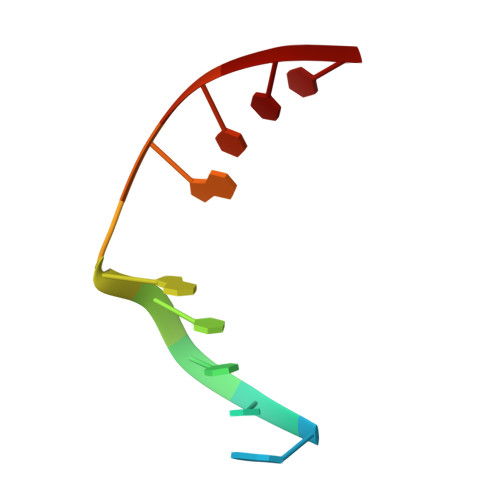
Structural Basis for Finding OG Lesions and Avoiding Undamaged G by the DNA Glycosylase MutY.
Russelburg, L.P., O'Shea Murray, V.L., Demir, M., Knutsen, K.R., Sehgal, S.L., Cao, S., David, S.S., Horvath, M.P.(2020) ACS Chem Biol 15: 93-102
- PubMed: 31829624
- DOI: https://doi.org/10.1021/acschembio.9b00639
- Primary Citation of Related Structures:
6Q0C, 6U7T - PubMed Abstract:
The adenine glycosylase MutY selectively initiates repair of OG:A lesions and, by comparison, avoids G:A mispairs. The ability to distinguish these closely related substrates relies on the C-terminal domain of MutY, which structurally resembles MutT. To understand the mechanism for substrate specificity, we crystallized MutY in complex with DNA containing G across from the high-affinity azaribose transition state analogue. Our structure shows that G is accommodated by the OG site and highlights the role of a serine residue in OG versus G discrimination. The functional significance of Ser308 and its neighboring residues was evaluated by mutational analysis, revealing the critical importance of a β loop in the C-terminal domain for mutation suppression in cells, and biochemical performance in vitro . This loop comprising residues Phe307, Ser308, and His309 ( Geobacillus stearothermophilus sequence positions) is conserved in MutY but absent in MutT and other DNA repair enzymes and may therefore serve as a MutY-specific target exploitable by chemical biological probes.
- School of Biological Sciences , University of Utah , 257 South 1400 East , Salt Lake City , Utah 84112 , United States.
Organizational Affiliation: