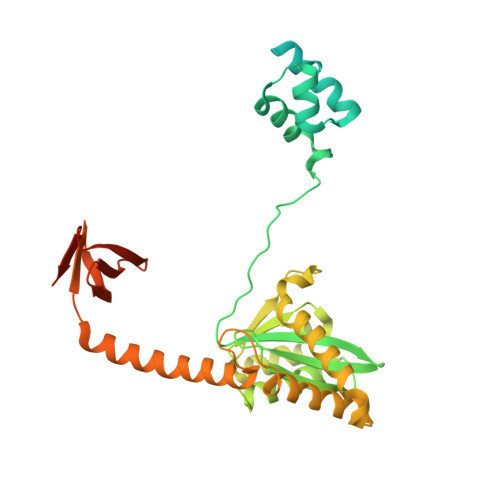
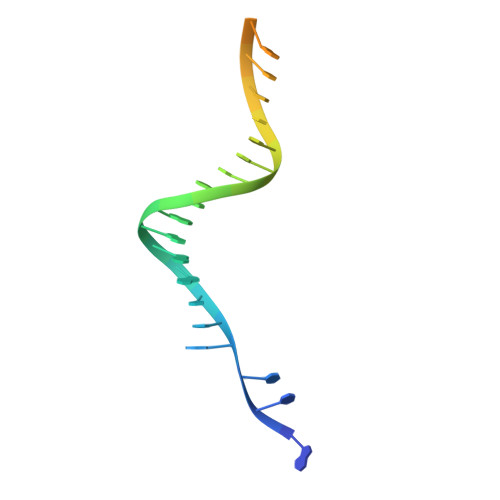
Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Passos, D.O., Li, M., Jozwik, I.K., Zhao, X.Z., Santos-Martins, D., Yang, R., Smith, S.J., Jeon, Y., Forli, S., Hughes, S.H., Burke Jr., T.R., Craigie, R., Lyumkis, D.(2020) Science 367: 810-814
- PubMed: 32001521
- DOI: https://doi.org/10.1126/science.aay8015
- Primary Citation of Related Structures:
6PUT, 6PUW, 6PUY, 6PUZ, 6V3K - PubMed Abstract:
The HIV intasome is a large nucleoprotein assembly that mediates the integration of a DNA copy of the viral genome into host chromatin. Intasomes are targeted by the latest generation of antiretroviral drugs, integrase strand-transfer inhibitors (INSTIs). Challenges associated with lentiviral intasome biochemistry have hindered high-resolution structural studies of how INSTIs bind to their native drug target. Here, we present high-resolution cryo-electron microscopy structures of HIV intasomes bound to the latest generation of INSTIs. These structures highlight how small changes in the integrase active site can have notable implications for drug binding and design and provide mechanistic insights into why a leading INSTI retains efficacy against a broad spectrum of drug-resistant variants. The data have implications for expanding effective treatments available for HIV-infected individuals.
- The Salk Institute for Biological Studies, Laboratory of Genetics, La Jolla, CA 92037, USA.
Organizational Affiliation: