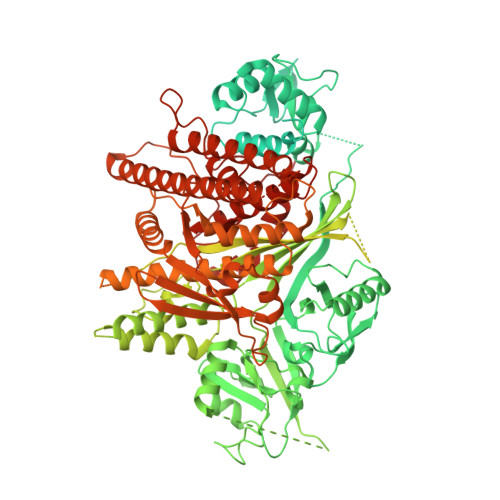
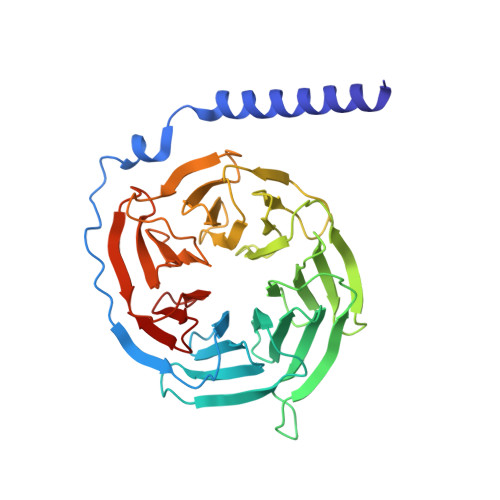
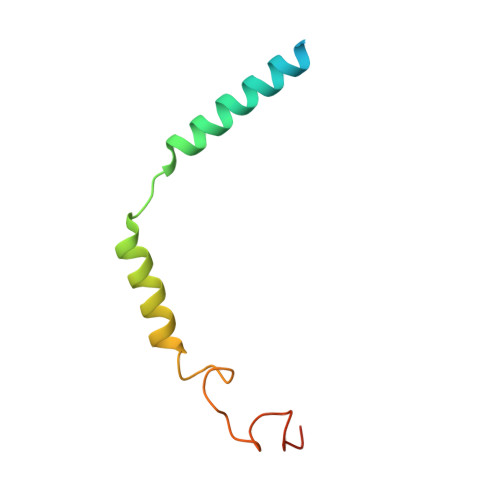
Cryo-electron microscopy structure and analysis of the P-Rex1-G beta gamma signaling scaffold.
Cash, J.N., Urata, S., Li, S., Ravala, S.K., Avramova, L.V., Shost, M.D., Gutkind, J.S., Tesmer, J.J.G., Cianfrocco, M.A.(2019) Sci Adv 5: eaax8855-eaax8855
- PubMed: 31663027
- DOI: https://doi.org/10.1126/sciadv.aax8855
- Primary Citation of Related Structures:
6PCV - PubMed Abstract:
PIP 3 -dependent Rac exchanger 1 (P-Rex1) is activated downstream of G protein-coupled receptors to promote neutrophil migration and metastasis. The structure of more than half of the enzyme and its regulatory G protein binding site are unknown. Our 3.2 Å cryo-EM structure of the P-Rex1-Gβγ complex reveals that the carboxyl-terminal half of P-Rex1 adopts a complex fold most similar to those of Legionella phosphoinositide phosphatases. Although catalytically inert, the domain coalesces with a DEP domain and two PDZ domains to form an extensive docking site for Gβγ. Hydrogen-deuterium exchange mass spectrometry suggests that Gβγ binding induces allosteric changes in P-Rex1, but functional assays indicate that membrane localization is also required for full activation. Thus, a multidomain assembly is key to the regulation of P-Rex1 by Gβγ and the formation of a membrane-localized scaffold optimized for recruitment of other signaling proteins such as PKA and PTEN.
- Department of Biological Chemistry & Life Sciences Institute, University of Michigan, Ann Arbor, MI, USA.
Organizational Affiliation: