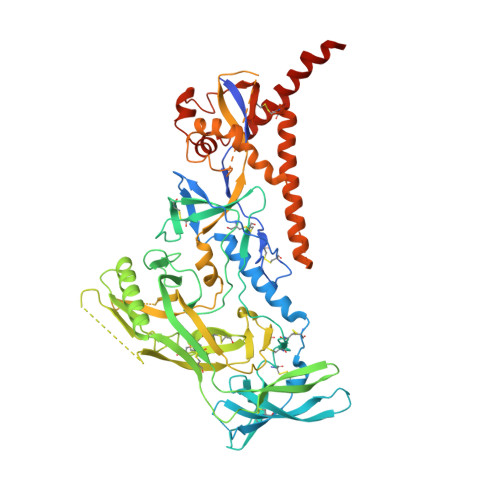
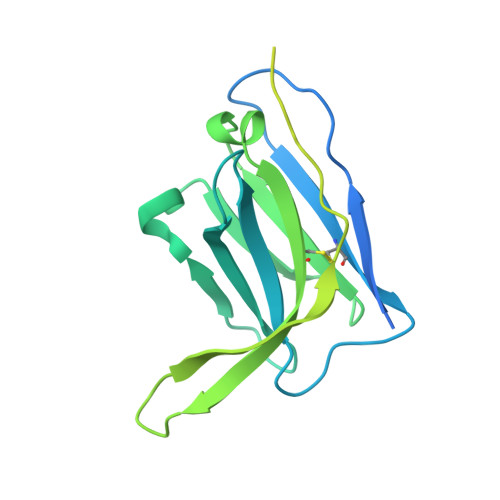
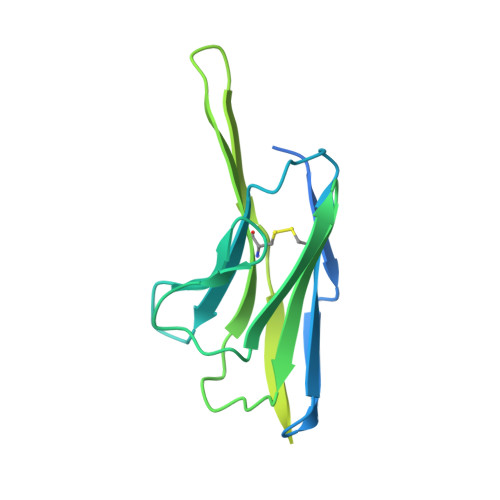
Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability.
Dubrovskaya, V., Tran, K., Ozorowski, G., Guenaga, J., Wilson, R., Bale, S., Cottrell, C.A., Turner, H.L., Seabright, G., O'Dell, S., Torres, J.L., Yang, L., Feng, Y., Leaman, D.P., Vazquez Bernat, N., Liban, T., Louder, M., McKee, K., Bailer, R.T., Movsesyan, A., Doria-Rose, N.A., Pancera, M., Karlsson Hedestam, G.B., Zwick, M.B., Crispin, M., Mascola, J.R., Ward, A.B., Wyatt, R.T.(2019) Immunity 51: 915-929.e7
- PubMed: 31732167
- DOI: https://doi.org/10.1016/j.immuni.2019.10.008
- Primary Citation of Related Structures:
6P62, 6P65, 6PEH - PubMed Abstract:
The elicitation of broadly neutralizing antibodies (bNAbs) against the HIV-1 envelope glycoprotein (Env) trimer remains a major vaccine challenge. Most cross-conserved protein determinants are occluded by self-N-glycan shielding, limiting B cell recognition of the underlying polypeptide surface. The exceptions to the contiguous glycan shield include the conserved receptor CD4 binding site (CD4bs) and glycoprotein (gp)41 elements proximal to the furin cleavage site. Accordingly, we performed heterologous trimer-liposome prime:boosting in rabbits to drive B cells specific for cross-conserved sites. To preferentially expose the CD4bs to B cells, we eliminated proximal N-glycans while maintaining the native-like state of the cleavage-independent NFL trimers, followed by gradual N-glycan restoration coupled with heterologous boosting. This approach successfully elicited CD4bs-directed, cross-neutralizing Abs, including one targeting a unique glycan-protein epitope and a bNAb (87% breadth) directed to the gp120:gp41 interface, both resolved by high-resolution cryoelectron microscopy. This study provides proof-of-principle immunogenicity toward eliciting bNAbs by vaccination.
- Department of Immunology and Microbiology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: