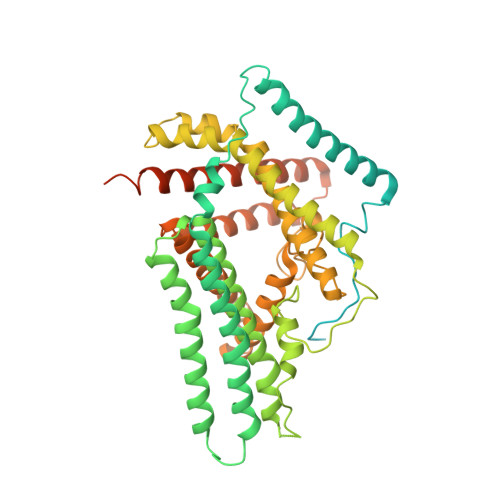
Structural basis for catalysis and substrate specificity of human ACAT1.
Qian, H., Zhao, X., Yan, R., Yao, X., Gao, S., Sun, X., Du, X., Yang, H., Wong, C.C.L., Yan, N.(2020) Nature 581: 333-338
- PubMed: 32433614
- DOI: https://doi.org/10.1038/s41586-020-2290-0
- Primary Citation of Related Structures:
6P2J, 6P2P - PubMed Abstract:
As members of the membrane-bound O-acyltransferase (MBOAT) enzyme family, acyl-coenzyme A:cholesterol acyltransferases (ACATs) catalyse the transfer of an acyl group from acyl-coenzyme A to cholesterol to generate cholesteryl ester, the primary form in which cholesterol is stored in cells and transported in plasma 1 . ACATs have gained attention as potential drug targets for the treatment of diseases such as atherosclerosis, Alzheimer's disease and cancer 2-7 . Here we present the cryo-electron microscopy structure of human ACAT1 as a dimer of dimers. Each protomer consists of nine transmembrane segments, which enclose a cytosolic tunnel and a transmembrane tunnel that converge at the predicted catalytic site. Evidence from structure-guided mutational analyses suggests that acyl-coenzyme A enters the active site through the cytosolic tunnel, whereas cholesterol may enter from the side through the transmembrane tunnel. This structural and biochemical characterization helps to rationalize the preference of ACAT1 for unsaturated acyl chains, and provides insight into the catalytic mechanism of enzymes within the MBOAT family 8 .
- Department of Molecular Biology, Princeton University, Princeton, NJ, USA. hongwuq@princeton.edu.
Organizational Affiliation: