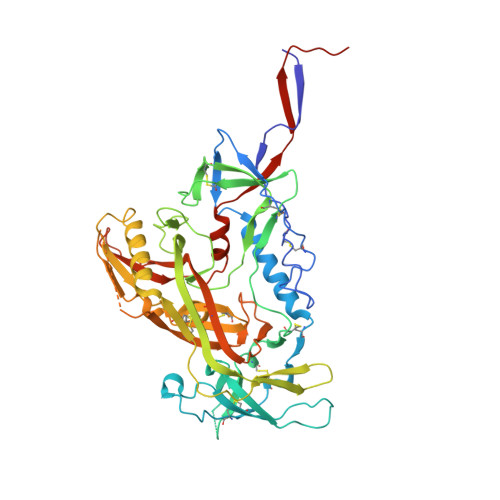
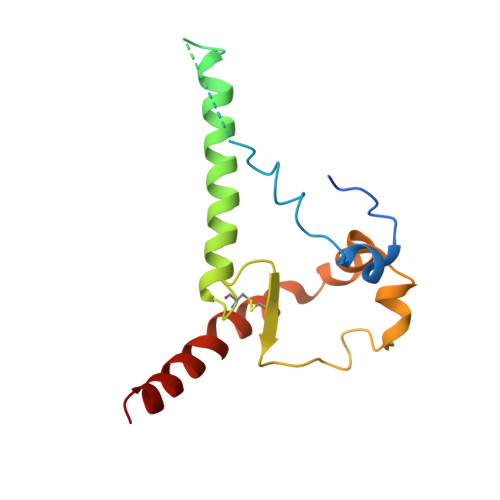
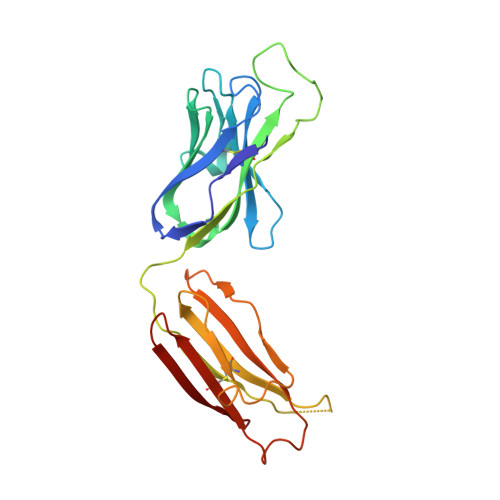
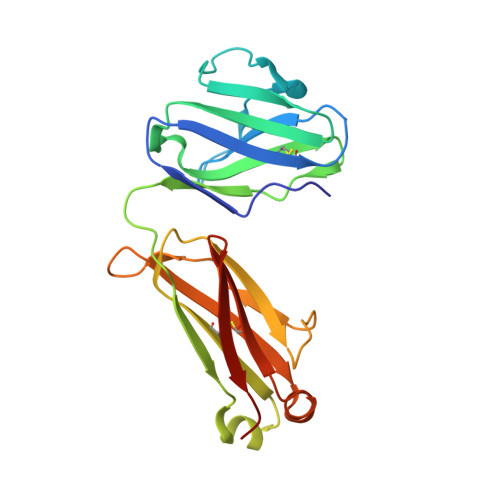
Near-Pan-neutralizing, Plasma Deconvoluted Antibody N49P6 Mimics Host Receptor CD4 in Its Quaternary Interactions with the HIV-1 Envelope Trimer.
Tolbert, W.D., Nguyen, D.N., Tehrani, Z.R., Sajadi, M.M., Pazgier, M.(2021) mBio : e0127421-e0127421
- PubMed: 34281393
- DOI: https://doi.org/10.1128/mBio.01274-21
- Primary Citation of Related Structures:
6OZ2, 6OZ4 - PubMed Abstract:
The first step in HIV-1 entry is the attachment of the envelope (Env) trimer to target cell CD4. As such, the CD4-binding site (CD4bs) remains one of the few universally accessible sites for antibodies (Abs). We recently described a method of isolating Abs directly from the circulating plasma and described a panel of broadly neutralizing Abs (bnAbs) from an HIV-1 "elite neutralizer" referred to as patient N49 (N49 Ab lineage [M. M. Sajadi, A. Dashti, Z. R. Tehrani, W. D. Tolbert, et al., Cell 173:1783-1795.e14, 2018, https://doi.org/10.1016/j.cell.2018.03.061]). Here, we describe the molecular details of antigen recognition by N49P6, an Ab of the N49 lineage that recapitulates most of the neutralization breadth and potency of the donor's plasma IgG. Our studies done in the context of monomeric and trimeric antigens indicate that N49P6 combines many characteristics of known CD4bs-specific bnAbs with features that are unique to the N49 Ab lineage to achieve its remarkable neutralization breadth. These include the omission of the CD4 Phe 43 cavity and dependence instead on interactions with highly conserved gp120 inner domain layer 3. Interestingly, when bound to BG505 SOSIP, N49P6 closely mimics the initial contact of host receptor CD4 to the adjacent promoter of the HIV-1 Env trimer to lock the trimer in the closed conformation. Altogether, N49P6 defines a new class of near-pan-neutralizing, plasma deconvoluted CD4bs Abs that we refer to as the N49P series. The details of the mechanisms of action of this new Ab class pave the way for the next generation of HIV-1 bnAbs that can be used as vaccine components of therapeutics. IMPORTANCE Binding to target cell CD4 is the first crucial step required for HIV-1 infection. Thus, the CD4-binding site (CD4bs) is one of the most accessible sites for antibodies (Abs). However, due to steric constraints, only a few Abs are capable of targeting this site. Here, we show that the exceptional neutralization breadth and potency of N49P6, a near-pan-neutralizing Ab targeting the CD4bs isolated from the plasma of an HIV-1 "elite neutralizer," patient N49, are due to its signature combination of more typical CD4bs Ab-binding characteristics with unique interactions with the highly conserved gp120 inner domain. In addition, we also present a structural analysis of N49P6 in complex with the BG505 SOSIP trimer to show that N49P6 exhibits remarkable breadth in part by mimicking CD4's quaternary interaction with the neighboring gp120 protomer. In its mode of antigen interaction, N49P6 is unique and represents a new class of CD4bs-specific bnAbs.
- Infectious Disease Division, Department of Medicine, Uniformed Services University of the Health Sciencesgrid.265436.0, Bethesda, Maryland, USA.
Organizational Affiliation: