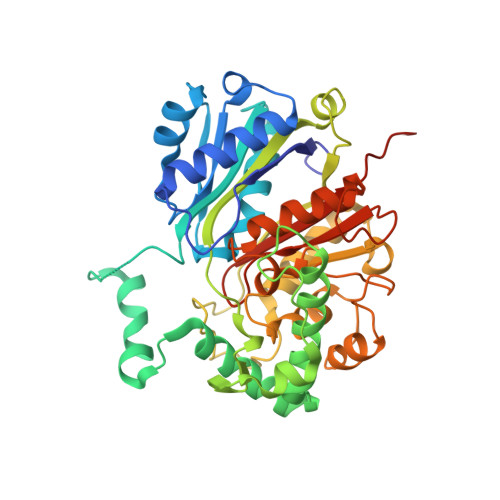
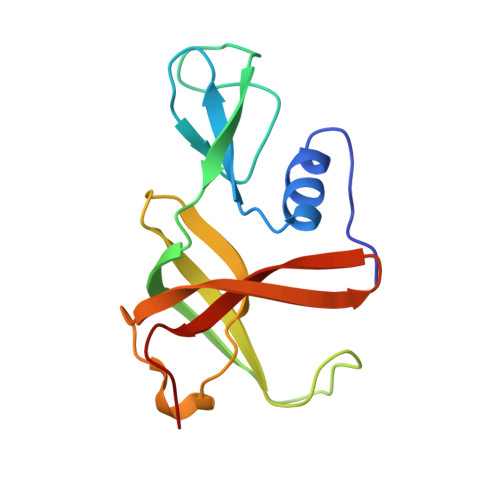
The steroid side-chain-cleaving aldolase Ltp2-ChsH2DUF35is a thiolase superfamily member with a radically repurposed active site.
Aggett, R., Mallette, E., Gilbert, S.E., Vachon, M.A., Schroeter, K.L., Kimber, M.S., Seah, S.Y.K.(2019) J Biological Chem 294: 11934-11943
- PubMed: 31209106
- DOI: https://doi.org/10.1074/jbc.RA119.008889
- Primary Citation of Related Structures:
6OK1 - PubMed Abstract:
An aldolase from the bile acid-degrading actinobacterium Thermomonospora curvata catalyzes the C-C bond cleavage of an isopropyl-CoA side chain from the D-ring of the steroid metabolite 17-hydroxy-3-oxo-4-pregnene-20-carboxyl-CoA (17-HOPC-CoA). Like its homolog from Mycobacterium tuberculosis , the T. curvata aldolase is a protein complex of Ltp2 with a DUF35 domain derived from the C-terminal domain of a hydratase (ChsH2 DUF35 ) that catalyzes the preceding step in the pathway. We determined the structure of the Ltp2-ChsH2 DUF35 complex at 1.7 Å resolution using zinc-single anomalous diffraction. The enzyme adopts an αββα organization, with the two Ltp2 protomers forming a central dimer, and the two ChsH2 DUF35 protomers being at the periphery. Docking experiments suggested that Ltp2 forms a tight complex with the hydratase but that each enzyme retains an independent CoA-binding site. Ltp2 adopted a fold similar to those in thiolases; however, instead of forming a deep tunnel, the Ltp2 active site formed an elongated cleft large enough to accommodate 17-HOPC-CoA. The active site lacked the two cysteines that served as the nucleophile and general base in thiolases and replaced a pair of oxyanion-hole histidine residues with Tyr-246 and Tyr-344. Phenylalanine replacement of either of these residues decreased aldolase catalytic activity at least 400-fold. On the basis of a 17-HOPC-CoA -docked model, we propose a catalytic mechanism where Tyr-294 acts as the general base abstracting a proton from the D-ring hydroxyl of 17-HOPC-CoA and Tyr-344 as the general acid that protonates the propionyl-CoA anion following C-C bond cleavage.
- Department of Molecular and Cellular Biology, University of Guelph, Guelph, Ontario N1G 5E9, Canada.
Organizational Affiliation: