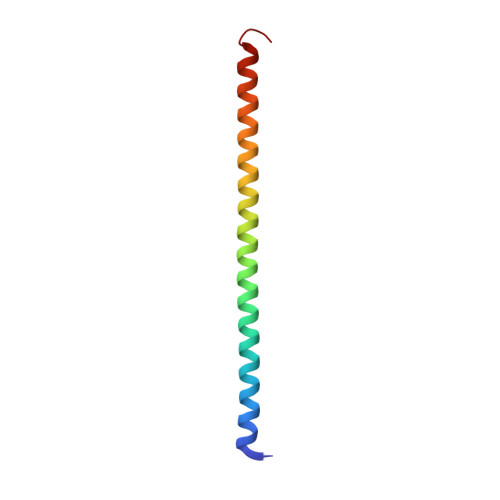Remarkable Rigidity of the Single alpha-Helical Domain of Myosin-VI As Revealed by NMR Spectroscopy.
Barnes, C.A., Shen, Y., Ying, J., Takagi, Y., Torchia, D.A., Sellers, J.R., Bax, A.(2019) J Am Chem Soc 141: 9004-9017
- PubMed: 31117653
- DOI: https://doi.org/10.1021/jacs.9b03116
- Primary Citation of Related Structures:
6OBI - PubMed Abstract:
Although the α-helix has long been recognized as an all-important element of secondary structure, it generally requires stabilization by tertiary interactions with other parts of a protein's structure. Highly charged single α-helical (SAH) domains, consisting of a high percentage (>75%) of Arg, Lys, and Glu residues, are exceptions to this rule but have been difficult to characterize structurally. Our study focuses on the 68-residue medial tail domain of myosin-VI, which is found to contain a highly ordered α-helical structure extending from Glu-6 to Lys-63. High hydrogen exchange protection factors (15-150), small (ca. 4 Hz) 3 J HNHα couplings, and a near-perfect fit to an ideal model α-helix for its residual dipolar couplings (RDCs), measured in a filamentous phage medium, support the high regularity of this helix. Remarkably, the hydrogen exchange rates are far more homogeneous than the protection factors derived from them, suggesting that for these transiently broken helices the intrinsic exchange rates derived from the amino acid sequence are not appropriate reference values. 15 N relaxation data indicate a very high degree of rotational diffusion anisotropy ( D ∥ / D ⊥ ≈ 7.6), consistent with the hydrodynamic behavior predicted for such a long, nearly straight α-helix. Alignment of the helix by a paramagnetic lanthanide ion attached to its N-terminal region shows a decrease in alignment as the distance from the tagging site increases. This decrease yields a precise measure for the persistence length of 224 ± 10 Å at 20 °C, supporting the idea that the role of the SAH helix is to act as an extension of the myosin-VI lever arm.