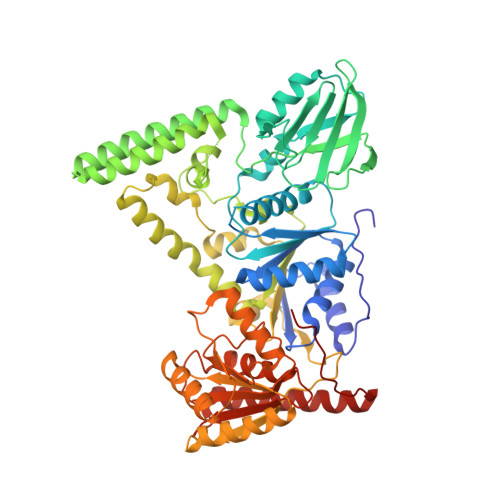
Mechanism of DNA Lesion Homing and Recognition by the Uvr Nucleotide Excision Repair System.
Lee, S.J., Sung, R.J., Verdine, G.L.(2019) Research (Wash D C) 2019: 5641746-5641746
- PubMed: 31549070
- DOI: https://doi.org/10.34133/2019/5641746
- Primary Citation of Related Structures:
6O8E, 6O8F, 6O8G, 6O8H - PubMed Abstract:
Nucleotide excision repair (NER) is an essential DNA repair system distinguished from other such systems by its extraordinary versatility. NER removes a wide variety of structurally dissimilar lesions having only their bulkiness in common. NER can also repair several less bulky nucleobase lesions, such as 8-oxoguanine. Thus, how a single DNA repair system distinguishes such a diverse array of structurally divergent lesions from undamaged DNA has been one of the great unsolved mysteries in the field of genome maintenance. Here we employ a synthetic crystallography approach to obtain crystal structures of the pivotal NER enzyme UvrB in complex with duplex DNA, trapped at the stage of lesion-recognition. These structures coupled with biochemical studies suggest that UvrB integrates the ATPase-dependent helicase/translocase and lesion-recognition activities. Our work also conclusively establishes the identity of the lesion-containing strand and provides a compelling insight to how UvrB recognizes a diverse array of DNA lesions.
- Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA.
Organizational Affiliation: