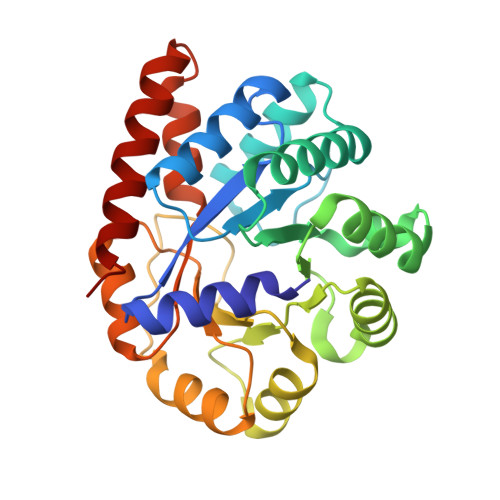
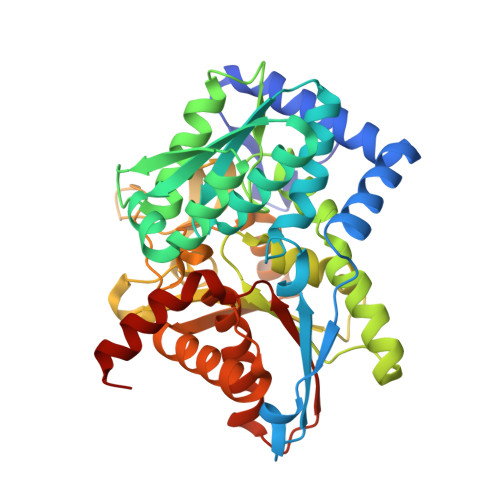
Crystal structure of Salmonella typhimurium Tryptophan Synthase mutant beta-Q114A with 2-({[4-(trifluoromethoxy)phenyl]sulfonyl}amino)ethyl dihydrogen phosphate (F9F) at the alpha-site, Cesium ion at the metal coordination site, and [3-hydroxy-2-methyl-5-phosphonooxymethyl-pyridin-4-ylmethyl]-serine (PLS) at the beta-site.
Hilario, E., Dunn, M.F., Mueller, L.J., Fan, L.To be published.