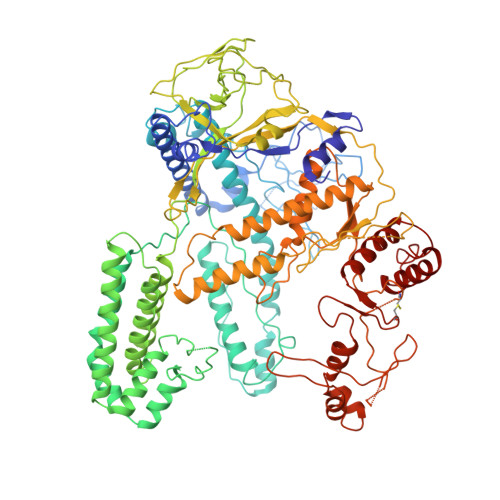
CasX enzymes comprise a distinct family of RNA-guided genome editors.
Liu, J.J., Orlova, N., Oakes, B.L., Ma, E., Spinner, H.B., Baney, K.L.M., Chuck, J., Tan, D., Knott, G.J., Harrington, L.B., Al-Shayeb, B., Wagner, A., Brotzmann, J., Staahl, B.T., Taylor, K.L., Desmarais, J., Nogales, E., Doudna, J.A.(2019) Nature 566: 218-223
- PubMed: 30718774
- DOI: https://doi.org/10.1038/s41586-019-0908-x
- Primary Citation of Related Structures:
6NY1, 6NY2, 6NY3 - PubMed Abstract:
The RNA-guided CRISPR-associated (Cas) proteins Cas9 and Cas12a provide adaptive immunity against invading nucleic acids, and function as powerful tools for genome editing in a wide range of organisms. Here we reveal the underlying mechanisms of a third, fundamentally distinct RNA-guided genome-editing platform named CRISPR-CasX, which uses unique structures for programmable double-stranded DNA binding and cleavage. Biochemical and in vivo data demonstrate that CasX is active for Escherichia coli and human genome modification. Eight cryo-electron microscopy structures of CasX in different states of assembly with its guide RNA and double-stranded DNA substrates reveal an extensive RNA scaffold and a domain required for DNA unwinding. These data demonstrate how CasX activity arose through convergent evolution to establish an enzyme family that is functionally separate from both Cas9 and Cas12a.
- Department of Molecular and Cell Biology, University of California, Berkeley, CA, USA.
Organizational Affiliation: