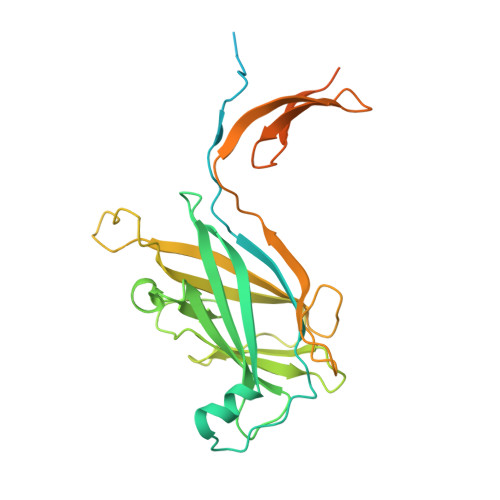
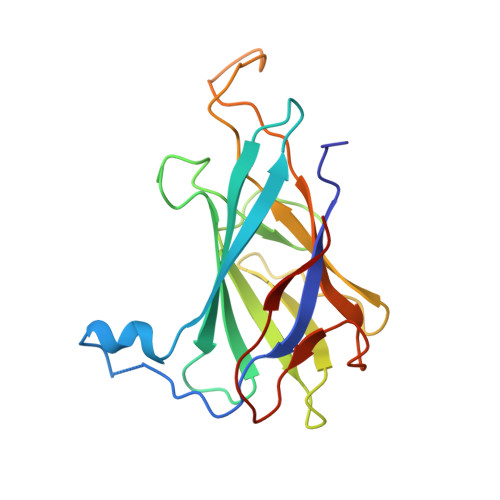
Structure and chemistry of lysinoalanine crosslinking in the spirochaete flagella hook.
Lynch, M.J., Miller, M., James, M., Zhang, S., Zhang, K., Li, C., Charon, N.W., Crane, B.R.(2019) Nat Chem Biol 15: 959-965
- PubMed: 31406373
- DOI: https://doi.org/10.1038/s41589-019-0341-3
- Primary Citation of Related Structures:
6NDT, 6NDV, 6NDW, 6NDX - PubMed Abstract:
The flagellar hook protein FlgE from spirochaete bacteria self-catalyzes the formation of an unusual inter-subunit lysinoalanine (Lal) crosslink that is critical for cell motility. Unlike other known examples of Lal biosynthesis, conserved cysteine and lysine residues in FlgE spontaneously react to form Lal without the involvement of additional enzymes. Oligomerization of FlgE via its D0 and Dc domains drives assembly of the crosslinking site at the D1-D2 domain interface. Structures of the FlgE D2 domain, dehydroalanine (DHA) intermediate and Lal crosslinked FlgE subunits reveal successive snapshots of the reaction. Cys178 flips from a buried configuration to release hydrogen sulfide (H 2 S/HS - ) and produce DHA. Interface residues provide hydrogen bonds to anchor the active site, facilitate β-elimination of Cys178 and polarize the peptide backbone to activate DHA for reaction with Lys165. Cysteine-reactive molecules accelerate DHA formation, whereas nucleophiles can intercept the DHA intermediate, thereby indicating a potential for Lal crosslink inhibitors to combat spirochaetal diseases.
- Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY, USA.
Organizational Affiliation: