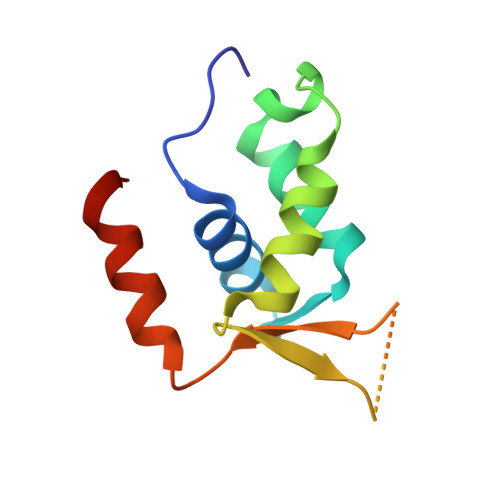
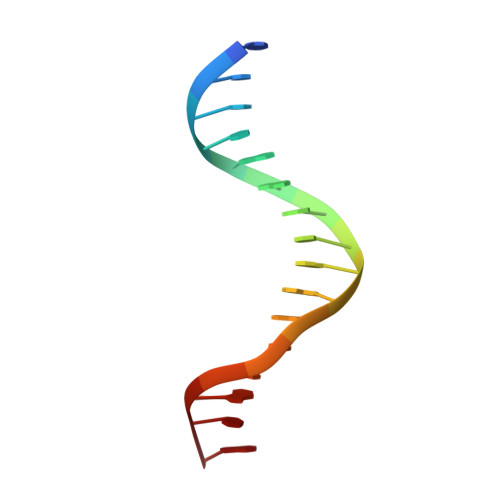
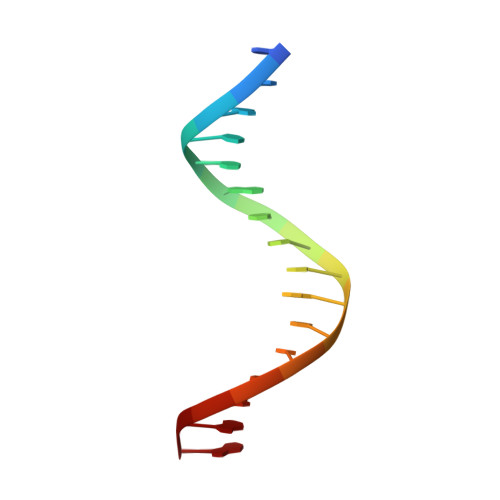
Bispecific Forkhead Transcription Factor FoxN3 Recognizes Two Distinct Motifs with Different DNA Shapes.
Rogers, J.M., Waters, C.T., Seegar, T.C.M., Jarrett, S.M., Hallworth, A.N., Blacklow, S.C., Bulyk, M.L.(2019) Mol Cell 74: 245
- PubMed: 30826165
- DOI: https://doi.org/10.1016/j.molcel.2019.01.019
- Primary Citation of Related Structures:
6NCE, 6NCM - PubMed Abstract:
Transcription factors (TFs) control gene expression by binding DNA recognition sites in genomic regulatory regions. Although most forkhead TFs recognize a canonical forkhead (FKH) motif, RYAAAYA, some forkheads recognize a completely different (FHL) motif, GACGC. Bispecific forkhead proteins recognize both motifs, but the molecular basis for bispecific DNA recognition is not understood. We present co-crystal structures of the FoxN3 DNA binding domain bound to the FKH and FHL sites, respectively. FoxN3 adopts a similar conformation to recognize both motifs, making contacts with different DNA bases using the same amino acids. However, the DNA structure is different in the two complexes. These structures reveal how a single TF binds two unrelated DNA sequences and the importance of DNA shape in the mechanism of bispecific recognition.
- Division of Genetics, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA; Committee on Higher Degrees in Biophysics, Harvard University, Cambridge, MA 02138, USA.
Organizational Affiliation: