Differential Misfolding Properties of Glaucoma-Associated Olfactomedin Domains from Humans and Mice.
Patterson-Orazem, A.C., Hill, S.E., Wang, Y., Dominic, I.M., Hall, C.K., Lieberman, R.L.(2019) Biochemistry 58: 1718-1727
- PubMed: 30802039
- DOI: https://doi.org/10.1021/acs.biochem.8b01309
- Primary Citation of Related Structures:
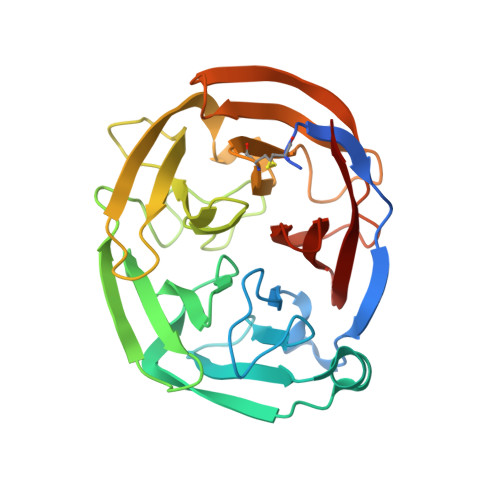
6NAX - PubMed Abstract:
Mutations in myocilin, predominantly within its olfactomedin (OLF) domain, are causative for the heritable form of open angle glaucoma in humans. Surprisingly, mice expressing Tyr423His mutant myocilin, corresponding to a severe glaucoma-causing mutation (Tyr437His) in human subjects, exhibit a weak, if any, glaucoma phenotype. To address possible protein-level discrepancies between mouse and human OLFs, which might lead to this outcome, biophysical properties of mouse OLF were characterized for comparison with those of human OLF. The 1.55 Å resolution crystal structure of mouse OLF reveals an asymmetric 5-bladed β-propeller that is nearly indistinguishable from previous structures of human OLF. Wild-type and selected mutant mouse OLFs mirror thermal stabilities of their human OLF counterparts, including characteristic stabilization in the presence of calcium. Mouse OLF forms thioflavin T-positive aggregates with a similar end-point morphology as human OLF, but amyloid aggregation kinetic rates of mouse OLF are faster than human OLF. Simulations and experiments support the interpretation that kinetics of mouse OLF are faster because of a decreased charge repulsion arising from more neutral surface electrostatics. Taken together, phenotypic differences observed in mouse and human studies of mutant myocilin could be a function of aggregation kinetics rates, which would alter the lifetime of putatively toxic protofibrillar intermediates.
- School of Chemistry & Biochemistry , Georgia Institute of Technology , Atlanta , Georgia 30332-0400 , United States.
Organizational Affiliation: