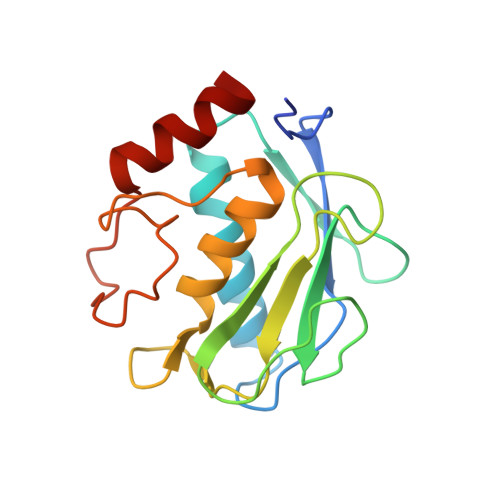
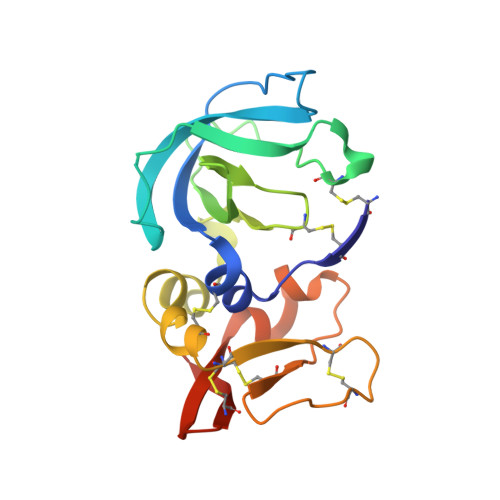
Directed evolution of the metalloproteinase inhibitor TIMP-1 reveals that its N- and C-terminal domains cooperate in matrix metalloproteinase recognition.
Raeeszadeh-Sarmazdeh, M., Greene, K.A., Sankaran, B., Downey, G.P., Radisky, D.C., Radisky, E.S.(2019) J Biological Chem 294: 9476-9488
- PubMed: 31040180
- DOI: https://doi.org/10.1074/jbc.RA119.008321
- Primary Citation of Related Structures:
6MAV, 6N9D - PubMed Abstract:
Tissue inhibitors of metalloproteinases (TIMPs) are natural inhibitors of matrix metalloproteinases (MMPs), enzymes that contribute to cancer and many inflammatory and degenerative diseases. The TIMP N-terminal domain binds and inhibits an MMP catalytic domain, but the role of the TIMP C-terminal domain in MMP inhibition is poorly understood. Here, we employed yeast surface display for directed evolution of full-length human TIMP-1 to develop MMP-3-targeting ultrabinders. By simultaneously incorporating diversity into both domains, we identified TIMP-1 variants that were up to 10-fold improved in binding MMP-3 compared with WT TIMP-1, with inhibition constants ( K i ) in the low picomolar range. Analysis of individual and paired mutations from the selected TIMP-1 variants revealed cooperative effects between distant residues located on the N- and C-terminal TIMP domains, positioned on opposite sides of the interaction interface with MMP-3. Crystal structures of MMP-3 complexes with TIMP-1 variants revealed conformational changes in TIMP-1 near the cooperative mutation sites. Affinity was strengthened by cinching of a reciprocal "tyrosine clasp" formed between the N-terminal domain of TIMP-1 and proximal MMP-3 interface and by changes in secondary structure within the TIMP-1 C-terminal domain that stabilize interdomain interactions and improve complementarity to MMP-3. Our protein engineering and structural studies provide critical insight into the cooperative function of TIMP domains and the significance of peripheral TIMP epitopes in MMP recognition. Our findings suggest new strategies to engineer TIMP proteins for therapeutic applications, and our directed evolution approach may also enable exploration of functional domain interactions in other protein systems.
- From the Department of Cancer Biology, Mayo Clinic, Jacksonville, Florida 32224.
Organizational Affiliation: