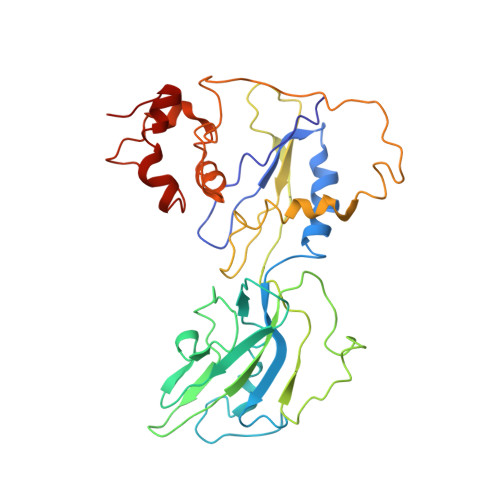
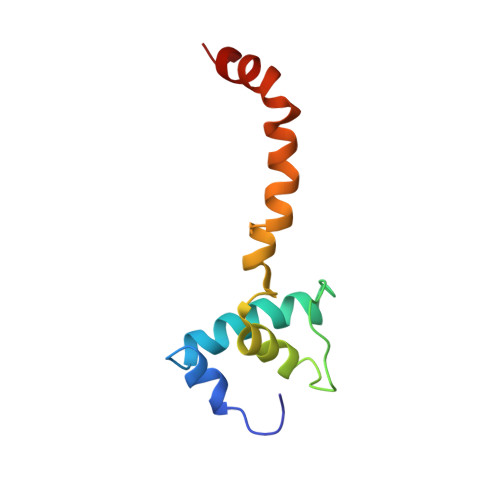
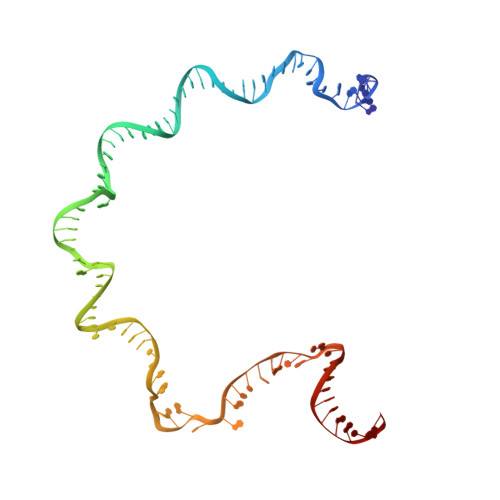
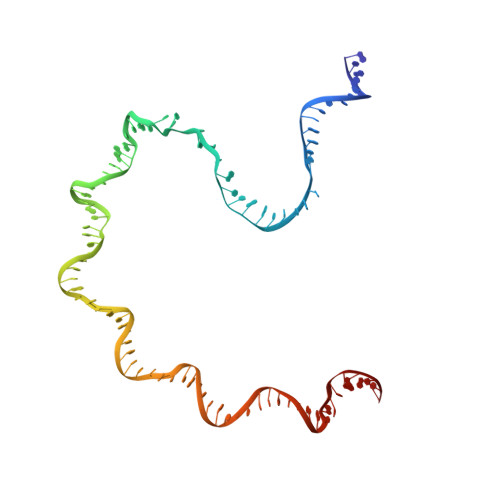
Interactions of Upstream and Downstream Promoter Regions with RNA Polymerase are Energetically Coupled and a Target of Regulation in Transcription Initiation
Sosa, R., Florez-Ariza, A., Diaz-Celis, C., Onoa, B., Cassago, A., de Oliveira, P.S.L., Portugal, R.V., Guerra, D.G., Bustamante, C.(2020) bioRxiv