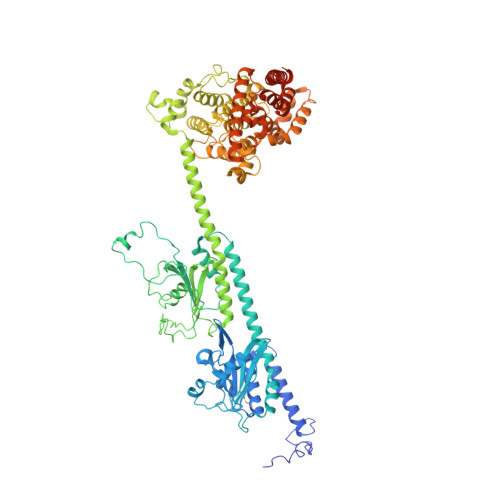
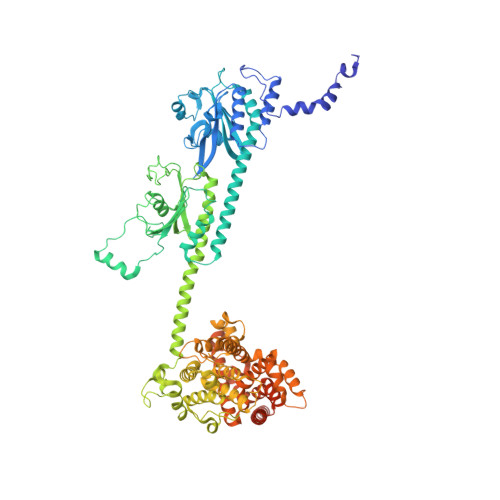
Cryo-EM structure of phosphodiesterase 6 reveals insights into the allosteric regulation of type I phosphodiesterases.
Gulati, S., Palczewski, K., Engel, A., Stahlberg, H., Kovacik, L.(2019) Sci Adv 5: eaav4322-eaav4322
- PubMed: 30820458
- DOI: https://doi.org/10.1126/sciadv.aav4322
- Primary Citation of Related Structures:
6MZB - PubMed Abstract:
Cyclic nucleotide phosphodiesterases (PDEs) work in conjunction with adenylate/guanylate cyclases to regulate the key second messengers of G protein-coupled receptor signaling. Previous attempts to determine the full-length structure of PDE family members at high-resolution have been hindered by structural flexibility, especially in their linker regions and N- and C-terminal ends. Therefore, most structure-activity relationship studies have so far focused on truncated and conserved catalytic domains rather than the regulatory domains that allosterically govern the activity of most PDEs. Here, we used single-particle cryo-electron microscopy to determine the structure of the full-length PDE6αβ2γ complex. The final density map resolved at 3.4 Å reveals several previously unseen structural features, including a coiled N-terminal domain and the interface of PDE6γ subunits with the PDE6αβ heterodimer. Comparison of the PDE6αβ2γ complex with the closed state of PDE2A sheds light on the conformational changes associated with the allosteric activation of type I PDEs.
- Gavin Herbert Eye Institute and the Department of Ophthalmology, University of California, Irvine, 829 Health Sciences Road, Irvine, CA 92617, USA.
Organizational Affiliation: