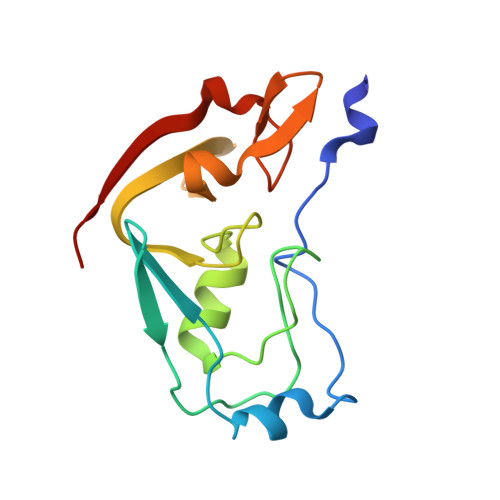
The structure of the cysteine-rich region from human histone-lysine N-methyltransferase EHMT2 (G9a).
Kerchner, K.M., Mou, T.C., Sun, Y., Rusnac, D.V., Sprang, S.R., Briknarova, K.(2021) J Struct Biol X 5: 100050-100050
- PubMed: 34278292
- DOI: https://doi.org/10.1016/j.yjsbx.2021.100050
- Primary Citation of Related Structures:
6MM1 - PubMed Abstract:
Euchromatic histone-lysine N-methyltransferase 1 (EHMT1; G9a-like protein; GLP) and euchromatic histone-lysine N-methyltransferase 2 (EHMT2; G9a) are protein lysine methyltransferases that regulate gene expression and are essential for development and the ability of organisms to change and adapt. In addition to ankyrin repeats and the catalytic SET domain, the EHMT proteins contain a unique cysteine-rich region (CRR) that mediates protein-protein interactions and recruitment of the methyltransferases to specific sites in chromatin. We have determined the structure of the CRR from human EHMT2 by X-ray crystallography and show that the CRR adopts an unusual compact fold with four bound zinc atoms. The structure consists of a RING domain preceded by a smaller zinc-binding motif and an N-terminal segment. The smaller zinc-binding motif straddles the N-terminal end of the RING domain, and the N-terminal segment runs in an extended conformation along one side of the structure and interacts with both the smaller zinc-binding motif and the RING domain. The interface between the N-terminal segment and the RING domain includes one of the zinc atoms. The RING domain is partially sequestered within the CRR and unlikely to function as a ubiquitin ligase.
- Department of Chemistry and Biochemistry, University of Montana, Missoula, MT 59812, USA.
Organizational Affiliation: