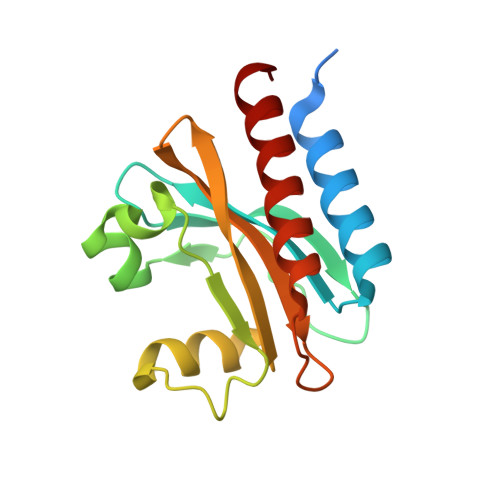
Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing.
Oliinyk, O.S., Shemetov, A.A., Pletnev, S., Shcherbakova, D.M., Verkhusha, V.V.(2019) Nat Commun 10: 279-279
- PubMed: 30655515
- DOI: https://doi.org/10.1038/s41467-018-08050-8
- Primary Citation of Related Structures:
6MGH - PubMed Abstract:
From a single domain of cyanobacteriochrome (CBCR) we developed a near-infrared (NIR) fluorescent protein (FP), termed miRFP670nano, with excitation at 645 nm and emission at 670 nm. This is the first CBCR-derived NIR FP evolved to efficiently bind endogenous biliverdin chromophore and brightly fluoresce in mammalian cells. miRFP670nano is a monomer with molecular weight of 17 kDa that is 2-fold smaller than bacterial phytochrome (BphP)-based NIR FPs and 1.6-fold smaller than GFP-like FPs. Crystal structure of the CBCR-based NIR FP with biliverdin reveals a molecular basis of its spectral and biochemical properties. Unlike BphP-derived NIR FPs, miRFP670nano is highly stable to denaturation and degradation and can be used as an internal protein tag. miRFP670nano is an effective FRET donor for red-shifted NIR FPs, enabling engineering NIR FRET biosensors spectrally compatible with GFP-like FPs and blue-green optogenetic tools. miRFP670nano unlocks a new source of diverse CBCR templates for NIR FPs.
- Medicum, Faculty of Medicine, University of Helsinki, 00290, Helsinki, Finland.
Organizational Affiliation: