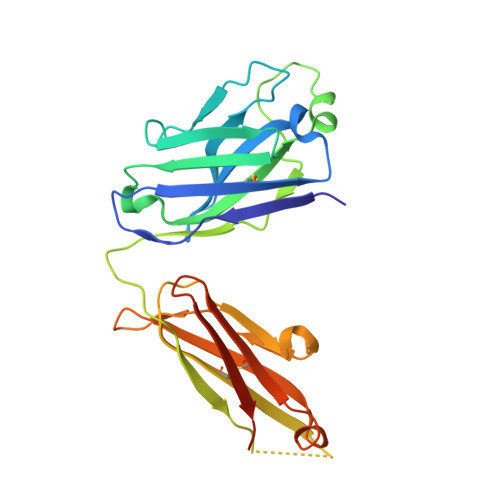
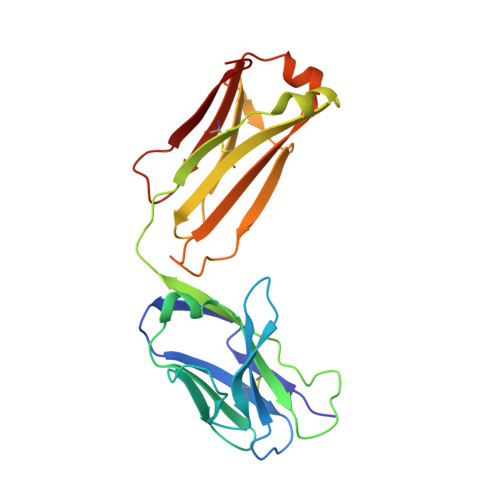
Structural basis of broad ebolavirus neutralization by a human survivor antibody.
West, B.R., Wec, A.Z., Moyer, C.L., Fusco, M.L., Ilinykh, P.A., Huang, K., Wirchnianski, A.S., James, R.M., Herbert, A.S., Hui, S., Goodwin, E., Howell, K.A., Kailasan, S., Aman, M.J., Walker, L.M., Dye, J.M., Bukreyev, A., Chandran, K., Saphire, E.O.(2019) Nat Struct Mol Biol 26: 204-212
- PubMed: 30833785
- DOI: https://doi.org/10.1038/s41594-019-0191-4
- Primary Citation of Related Structures:
6MAM - PubMed Abstract:
The structural features that govern broad-spectrum activity of broadly neutralizing anti-ebolavirus antibodies (Abs) outside of the internal fusion loop epitope are currently unknown. Here we describe the structure of a broadly neutralizing human monoclonal Ab (mAb), ADI-15946, which was identified in a human survivor of the 2013-2016 outbreak. The crystal structure of ADI-15946 in complex with cleaved Ebola virus glycoprotein (EBOV GP CL ) reveals that binding of the mAb structurally mimics the conserved interaction between the EBOV GP core and its glycan cap β17-β18 loop to inhibit infection. Both endosomal proteolysis of EBOV GP and binding of mAb FVM09 displace this loop, thereby increasing exposure of ADI-15946's conserved epitope and enhancing neutralization. Our work also mapped the paratope of ADI-15946, thereby explaining reduced activity against Sudan virus, which enabled rational, structure-guided engineering to enhance binding and neutralization of Sudan virus while retaining the parental activity against EBOV and Bundibugyo virus.
- Department of Immunology and Microbiology, The Scripps Research Institute, La Jolla, CA, USA.
Organizational Affiliation: