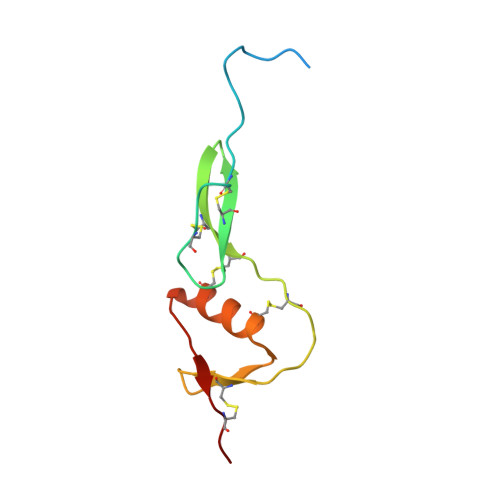
Crystal structure of the WFIKKN2 follistatin domain reveals insight into how it inhibits growth differentiation factor 8 (GDF8) and GDF11.
McCoy, J.C., Walker, R.G., Murray, N.H., Thompson, T.B.(2019) J Biological Chem 294: 6333-6343
- PubMed: 30814254
- DOI: https://doi.org/10.1074/jbc.RA118.005831
- Primary Citation of Related Structures:
6MAA - PubMed Abstract:
Growth differentiation factor 8 (GDF8; also known as myostatin) and GDF11 are closely related members of the transforming growth factor β (TGF-β) family. GDF8 strongly and negatively regulates skeletal muscle growth, and GDF11 has been implicated in various age-related pathologies such as cardiac hypertrophy. GDF8 and GDF11 signaling activities are controlled by the extracellular protein antagonists follistatin; follistatin-like 3 (FSTL3); and WAP, follistatin/kazal, immunoglobulin, Kunitz, and netrin domain-containing (WFIKKN). All of these proteins contain a follistatin domain (FSD) important for ligand binding and antagonism. Here, we investigated the structure and function of the FSD from murine WFIKKN2 and compared it with the FSDs of follistatin and FSTL3. Using native gel shift and surface plasmon resonance analyses, we determined that the WFIKKN2 FSD can interact with both GDF8 and GDF11 and block their interactions with the type II receptor activin A receptor type 2B (ActRIIB). Further, we solved the crystal structure of the WFIKKN2 FSD to 1.39 Å resolution and identified surface-exposed residues that, when substituted with alanine, reduce antagonism of GDF8 in full-length WFIKKN2. Comparison of the WFIKKN2 FSD with those of follistatin and FSTL3 revealed differences in both the FSD structure and position of residues within the domain that are important for ligand antagonism. Taken together, our results indicate that both WFIKKN and follistatin utilize their FSDs to block the type II receptor but do so via different binding interactions.
- From the Department of Molecular Genetics, Biochemistry, and Microbiology, College of Medicine, University of Cincinnati, Cincinnati, Ohio 45267.
Organizational Affiliation: