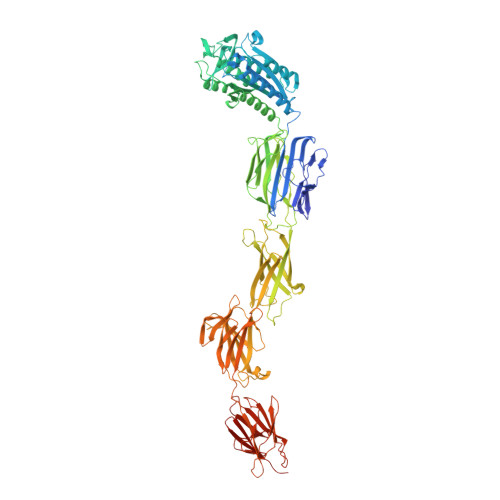
Crystal structure of lactobacillar SpaC reveals an atypical five-domain pilus tip adhesin: Exposing its substrate-binding and assembly in SpaCBA pili.
Kant, A., Palva, A., von Ossowski, I., Krishnan, V.(2020) J Struct Biol 211: 107571-107571
- PubMed: 32653644
- DOI: https://doi.org/10.1016/j.jsb.2020.107571
- Primary Citation of Related Structures:
6M3Y, 6M48, 6M7C, 7BVX - PubMed Abstract:
Adhesion to cell surfaces is an essential and early prerequisite for successful host colonization by bacteria, and in most instances involves the specificities of various adhesins. Among bacterial Gram-positives, some genera and species mediate attachment to host cells by using long non-flagellar appendages called sortase-dependent pili. A case in point is the beneficial Lactobacillus rhamnosus GG gut-adapted strain that produces the so-called SpaCBA pilus, a structure noted for its promiscuous binding to intestinal mucus and collagen. Structurally, SpaCBA pili are heteropolymers of three different pilin-protein subunits, each with its own location and function in the pilus: backbone SpaA for length, basal SpaB for anchoring, and tip SpaC for adhesion. Previously, we solved the SpaA tertiary structure by X-ray crystallography and also reported on the crystallization of SpaB and SpaC. Here, we reveal the full-length high-resolution (1.9 Å) crystal structure of SpaC, a first for a sortase-dependent pilus-bearing commensal. The SpaC structure, unlike the representative four-domain architecture of other Gram-positive tip pilins, espouses an atypically longer five-domain arrangement that includes N-terminal 'binding' and C-terminal 'stalk' regions of two and three domains, respectively. With the prospect of establishing new mechanistic insights, we provide a structural basis for the multi-substrate binding nature of SpaC, as well as a structural model that reconciles its exclusive localization at the SpaCBA pilus tip.
- Laboratory of Structural Microbiology, Regional Centre for Biotechnology, NCR Biotech Science Cluster, Faridabad 122016, India; Department of Biotechnology, Manipal University, Karnataka 576104, India.
Organizational Affiliation: