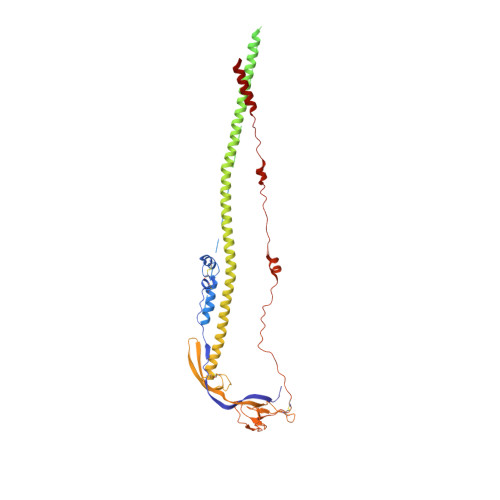
Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein.
Fan, X., Cao, D., Kong, L., Zhang, X.(2020) Nat Commun 11: 3618-3618
- PubMed: 32681106
- DOI: https://doi.org/10.1038/s41467-020-17371-6
- Primary Citation of Related Structures:
6M3W - PubMed Abstract:
Global emergencies caused by the severe acute respiratory syndrome coronavirus (SARS-CoV), Middle-East respiratory syndrome coronavirus (MERS-CoV) and SARS-CoV-2 significantly endanger human health. The spike (S) glycoprotein is the key antigen and its conserved S2 subunit contributes to viral entry by mediating host-viral membrane fusion. However, structural information of the post-fusion S2 from these highly pathogenic human-infecting coronaviruses is still lacking. We used single-particle cryo-electron microscopy to show that the post-fusion SARS-CoV S2 forms a further rotated HR1-HR2 six-helix bundle and a tightly bound linker region upstream of the HR2 motif. The structures of pre- and post-fusion SARS-CoV S glycoprotein dramatically differ, resembling that of the Mouse hepatitis virus (MHV) and other class I viral fusion proteins. This structure suggests potential targets for the development of vaccines and therapies against a wide range of SARS-like coronaviruses.
- National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, 100101, Beijing, China.
Organizational Affiliation: