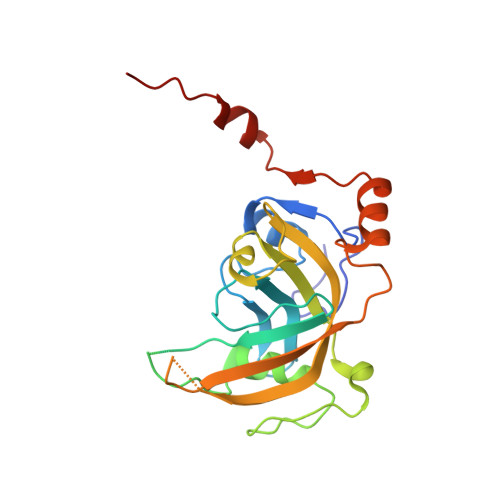
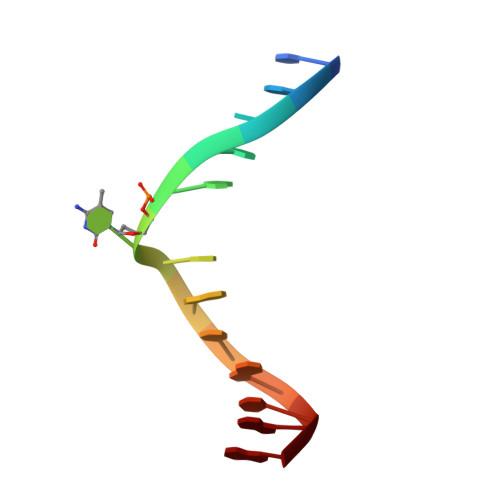
Mechanistic insights into recognition of symmetric methylated cytosines in CpG and non-CpG DNA by UHRF1 SRA.
Abhishek, S., Nakarakanti, N.K., Deeksha, W., Rajakumara, E.(2021) Int J Biol Macromol 170: 514-522
- PubMed: 33359809
- DOI: https://doi.org/10.1016/j.ijbiomac.2020.12.149
- Primary Citation of Related Structures:
6M2V - PubMed Abstract:
Non-CpG DNA methylation (non-mCpG) is enriched in the genome of brain neurons and germline cells in mammals. Accumulation of non-mCpG during postnatal brain development correlates with gene regulation and inactivation of distal regulatory elements. Recently, UHRF1 has been found to contribute to de novo non-CpG methylation, however, whether UHRF1 could recognize non-mCpG is unknown. Here, we have demonstrated through calorimetric measurements that the UHRF1 SRA can recognize mCpH and fully-mCpHpG, types of non-mCpG. Our ITC binding studies endorse the preferential reading of hemi-mCpG by UHRF1 SRA and also show 6-fold weaker binding for fully-mCpG than hemi-mCpG. Despite presence of symmetrical (5-methyl cytosine) 5mCs, stoichiometry of 1:1 for UHRF1 SRA binding to fully-mCpG indicates that UHRF1 SRA may not form a stable complex with fully-mCpG DNA. Contrarily, UHRF1 SRA recognizes fully-mCpHpG with a stoichiometry of 2:1 protein to DNA duplex with binding affinity higher than fully-mCpG. Our crystal structure of UHRF1 SRA bound to fully-mCpHpG DNA reveals dual flip-out mechanism of 5mC recognition. Metadynamics studies corroborates with ITC data that UHRF1 SRA could not form a stable complex with fully-mCpG DNA. Altogether, this study demonstrates that UHRF1 SRA recognizes non-mCpG DNA and exhibits contrasting mechanisms for hemi-mCpG and fully-mCpHpG DNA recognition.
- Macromolecular Structural Biology Lab, Department of Biotechnology, Indian Institute of Technology Hyderabad, Kandi, Sangareddy, Telangana 502285, India.
Organizational Affiliation: