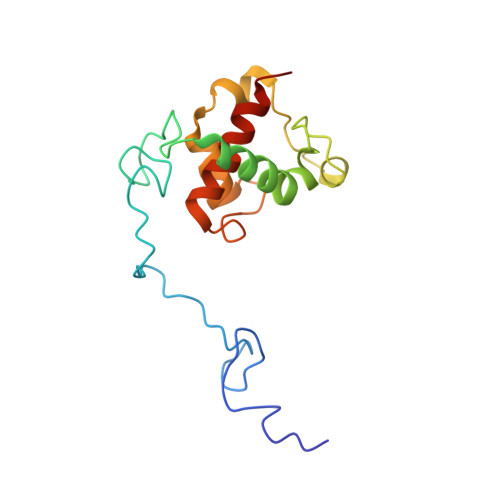
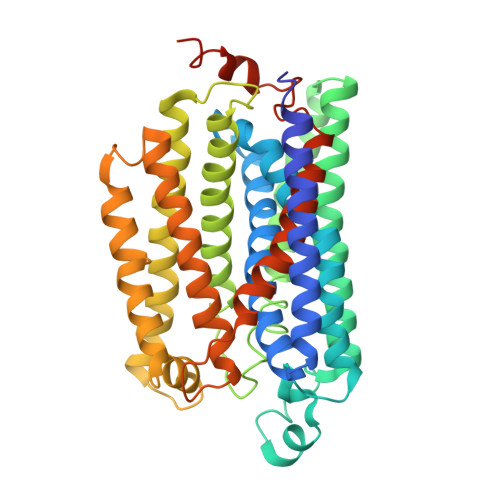
Cryo-EM structures of the air-oxidized and dithionite-reduced photosynthetic alternative complex III from Roseiflexus castenholzii .
Shi, Y., Xin, Y., Wang, C., Blankenship, R.E., Sun, F., Xu, X.(2020) Sci Adv 6: eaba2739-eaba2739
- PubMed: 32832681
- DOI: https://doi.org/10.1126/sciadv.aba2739
- Primary Citation of Related Structures:
6LOD, 6LOE - PubMed Abstract:
Alternative complex III (ACIII) is a multisubunit quinol:electron acceptor oxidoreductase that couples quinol oxidation with transmembrane proton translocation in both the respiratory and photosynthetic electron transport chains of bacteria. The coupling mechanism, however, is poorly understood. Here, we report the cryo-EM structures of air-oxidized and dithionite-reduced ACIII from the photosynthetic bacterium Roseiflexus castenholzii at 3.3- and 3.5-Å resolution, respectively. We identified a menaquinol binding pocket and an electron transfer wire comprising six hemes and four iron-sulfur clusters that is capable of transferring electrons to periplasmic acceptors. We detected a proton translocation passage in which three strictly conserved, mid-passage residues are likely essential for coupling the redox-driven proton translocation across the membrane. These results allow us to propose a previously unrecognized coupling mechanism that links the respiratory and photosynthetic functions of ACIII. This study provides a structural basis for further investigation of the energy transformation mechanisms in bacterial photosynthesis and respiration.
- National Laboratory of Biomacromolecules, National Center of Protein Science-Beijing, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: